PAGA
Last updated: 2019-04-03
workflowr checks: (Click a bullet for more information)-
✔ R Markdown file: up-to-date
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
-
✔ Environment: empty
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
-
✔ Seed:
set.seed(20190110)The command
set.seed(20190110)was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible. -
✔ Session information: recorded
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
-
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility. The version displayed above was the version of the Git repository at the time these results were generated.✔ Repository version: 5870e6e
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can usewflow_publishorwflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.Ignored files: Ignored: .DS_Store Ignored: .Rhistory Ignored: .Rproj.user/ Ignored: ._.DS_Store Ignored: analysis/cache/ Ignored: build-logs/ Ignored: data/alevin/ Ignored: data/cellranger/ Ignored: data/processed/ Ignored: data/published/ Ignored: output/.DS_Store Ignored: output/._.DS_Store Ignored: output/03-clustering/selected_genes.csv.zip Ignored: output/04-marker-genes/de_genes.csv.zip Ignored: packrat/.DS_Store Ignored: packrat/._.DS_Store Ignored: packrat/lib-R/ Ignored: packrat/lib-ext/ Ignored: packrat/lib/ Ignored: packrat/src/ Untracked files: Untracked: DGEList.Rds Untracked: output/90-methods/package-versions.json Untracked: scripts/build.pbs Unstaged changes: Modified: analysis/_site.yml Modified: output/01-preprocessing/droplet-selection.pdf Modified: output/01-preprocessing/parameters.json Modified: output/01-preprocessing/selection-comparison.pdf Modified: output/01B-alevin/alevin-comparison.pdf Modified: output/01B-alevin/parameters.json Modified: output/02-quality-control/qc-thresholds.pdf Modified: output/02-quality-control/qc-validation.pdf Modified: output/03-clustering/cluster-comparison.pdf Modified: output/03-clustering/cluster-validation.pdf Modified: output/04-marker-genes/de-results.pdf
Expand here to see past versions:
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| html | 33ac14f | Luke Zappia | 2019-03-20 | Tidy up website |
| html | ae75188 | Luke Zappia | 2019-03-06 | Revise figures after proofread |
| html | 2693e97 | Luke Zappia | 2019-03-05 | Add methods page |
| html | fefdd07 | Luke Zappia | 2019-02-28 | Add PAGA figure |
| Rmd | dd11830 | Luke Zappia | 2019-02-12 | Add PAGA text |
| html | dd11830 | Luke Zappia | 2019-02-12 | Add PAGA text |
| html | ee99ad1 | Luke Zappia | 2019-02-10 | Add PAGA |
# scRNA-seq
library("SingleCellExperiment")
# System
library("processx")
# Plotting
library("cowplot")
# Presentation
library("knitr")
# Tidyverse
library("tidyverse")source(here::here("R/output.R"))
source(here::here("R/plotting.R"))clust_path <- here::here("data/processed/03-clustered.Rds")
paga_path <- here::here("scripts/paga.py")Introduction
In this document we are going to explore the relationship between our clusters using partition-based graph abstraction (PAGA). PAGA builds a graph where each node is a cluster and the edges represent connectivity between clusters. This can give us an overview of the dataset and the relationships between cell types.
if (file.exists(clust_path)) {
sce <- read_rds(clust_path)
} else {
stop("Clustered dataset is missing. ",
"Please run '03-clustering.Rmd' first.",
call. = FALSE)
}PAGA
PAGA is available as part of the scanpy Python package. The PAGA analysis is performed using the following Python script.
cat(readLines(paga_path), sep = "\n")#!/usr/bin/env python3
import os
import json
import numpy
import pandas as pd
import loompy
import anndata
import scanpy as sc
# Show errors (0), warnings (1), info (2) and hints (3)
sc.settings.verbosity = 3
print('Reading clustering parameters...')
params_path = 'output/03-clustering/parameters.json'
with open(params_path) as params_file:
params = json.load(params_file)
for param in params:
if param['Parameter'][0] == 'n_pcs':
n_pcs = param['Value'][0]
if param['Parameter'][0] == 'knn':
knn = param['Value'][0]
print('Reading Loom file...')
loom_path = 'data/processed/03-clustered-sel.loom'
col_names = ['Cell', 'Barcode', 'Dataset', 'Sample', 'Cluster']
obs = dict()
with loompy.connect(loom_path) as loom_con:
X = loom_con.layers[''][()].T
for col in col_names:
obs[col] = loom_con.col_attrs[col]
print('Converting to AnnData...')
adata = anndata.AnnData(X=X, obs=obs)
print('Calculating PCs...')
sc.tl.pca(adata, svd_solver='arpack')
print('Calculating neighbour graph...')
sc.pp.neighbors(adata, n_neighbors=30, n_pcs=15)
print('Perfoming PAGA...')
sc.tl.paga(adata, groups='Cluster')
print('Calculating cluster graph layout...')
sc.pl.paga(adata, plot=False)
print('Calculating cell graph layout...')
sc.tl.draw_graph(adata, init_pos='paga')
out_dir = 'output/05-paga'
if not os.path.exists(out_dir):
os.makedirs(out_dir)
def con2edges(con, names=None, sparse=True):
print('Converting connectivity matrix to edges...')
n = con.shape[0]
edges = pd.DataFrame(columns=['From', 'To', 'Connectivity'])
for i in range(n):
for j in range(i + 1, n):
if names is not None:
fr = names[i]
to = names[j]
else:
fr = str(i)
to = str(j)
connectivity = con[i, j]
if sparse and connectivity == 0:
continue
entry = {'From' : fr, 'To' : to,
'Connectivity' : con[i, j]}
edges = edges.append(entry, ignore_index=True)
return edges
print('Outputting cluster edges...')
clust_con = adata.uns['paga']['connectivities'].toarray()
clust_edges = con2edges(clust_con)
clust_edges.to_csv(os.path.join(out_dir, 'cluster_edges.csv'),
index=False)
print('Outputting cluster tree edges...')
clust_tree_con = adata.uns['paga']['connectivities_tree'].toarray()
clust_tree_edges = con2edges(clust_tree_con)
clust_tree_edges.to_csv(os.path.join(out_dir, 'cluster_tree_edges.csv'),
index=False)
print('Outputting cluster embedding...')
clust_embedding = pd.DataFrame(adata.uns['paga']['pos'], columns=['X', 'Y'])
clust_embedding['Cluster'] = range(clust_embedding.shape[0])
clust_embedding = clust_embedding[['Cluster', 'X', 'Y']]
clust_embedding.to_csv(os.path.join(out_dir, 'cluster_embedding.csv'),
index=False)
print('Outputting cell edges...')
cells = adata.obs['Cell']
cell_con = adata.uns['neighbors']['connectivities']
cell_edges = pd.DataFrame(columns=['From', 'To', 'Connectivity'])
n_rows = len(cell_con.indptr)
for i in range(len(cell_con.indptr) - 1):
row_ind = cell_con.indices[cell_con.indptr[i]:cell_con.indptr[i + 1]]
print(f'\r\tRow {i} of {n_rows}', end='')
for k, j in enumerate(row_ind):
if j > i:
con = cell_con.data[cell_con.indptr[i] + k]
fr = cells[i]
to = cells[j]
entry = {'From' : fr, 'To' : to, 'Connectivity' : con}
cell_edges = cell_edges.append(entry, ignore_index=True)
print('\n')
cell_edges.to_csv(os.path.join(out_dir, 'cell_edges.csv'),
index=False)
print('Outputting cell embedding...')
x = adata.obsm['X_draw_graph_fa'][:, 0]
y = adata.obsm['X_draw_graph_fa'][:, 1]
cell_embedding = pd.DataFrame({'Cell' : cells, 'X' : x, 'Y' : y})
cell_embedding.to_csv(os.path.join(out_dir, 'cell_embedding.csv'),
index=False)
print('Done!')paga_out <- run(here::here("scripts/run_paga.sh"))We then load the PAGA results and visualise them here.
clust_embedding <- read_csv(
here::here("output/05-paga/cluster_embedding.csv"),
col_types = cols(
.default = col_double()
)
) %>%
mutate(Size = as.numeric(table(colData(sce)$Cluster))) %>%
mutate(Cluster = factor(Cluster, levels = levels(colData(sce)$Cluster)))
clust_edges <- read_csv(
here::here("output/05-paga/cluster_edges.csv"),
col_types = cols(
.default = col_double()
)
) %>%
mutate(To = factor(To, levels = levels(colData(sce)$Cluster)),
From = factor(From, levels = levels(colData(sce)$Cluster))) %>%
left_join(clust_embedding, by = c("From" = "Cluster")) %>%
rename(FromX = X, FromY = Y) %>%
select(-Size) %>%
left_join(clust_embedding, by = c("To" = "Cluster")) %>%
rename(ToX = X, ToY = Y) %>%
select(-Size)
cell_embedding <- read_csv(
here::here("output/05-paga/cell_embedding.csv"),
col_types = cols(
.default = col_double(),
Cell = col_character()
)
) %>%
mutate(Cluster = colData(sce)$Cluster)
cell_edges <- read_csv(
here::here("output/05-paga/cell_edges.csv"),
col_types = cols(
.default = col_double(),
From = col_character(),
To = col_character()
)
) %>%
left_join(cell_embedding, by = c("From" = "Cell")) %>%
rename(FromX = X, FromY = Y) %>%
select(-Cluster) %>%
left_join(cell_embedding, by = c("To" = "Cell")) %>%
rename(ToX = X, ToY = Y) %>%
select(-Cluster)Cluster graph
The PAGA graph shows the relationship between clusters. Here we have use a force directed graph layout in order to visualise it. PAGA calculates the connectivity between each pair of clusters but we need to apply some threshold to that to select the meaningful edges.
Thresholds
src_list <- lapply(seq(0, 0.9, 0.1), function(thresh) {
src <- c(
"### Con {{thresh}} {.unnumbered}",
"```{r clust-paga-{{thresh}}}",
"plotPAGAClustGraph(clust_embedding, clust_edges, thresh = {{thresh}})",
"```",
""
)
knit_expand(text = src)
})
out <- knit_child(text = unlist(src_list), options = list(cache = FALSE))Con 0
plotPAGAClustGraph(clust_embedding, clust_edges, thresh = 0)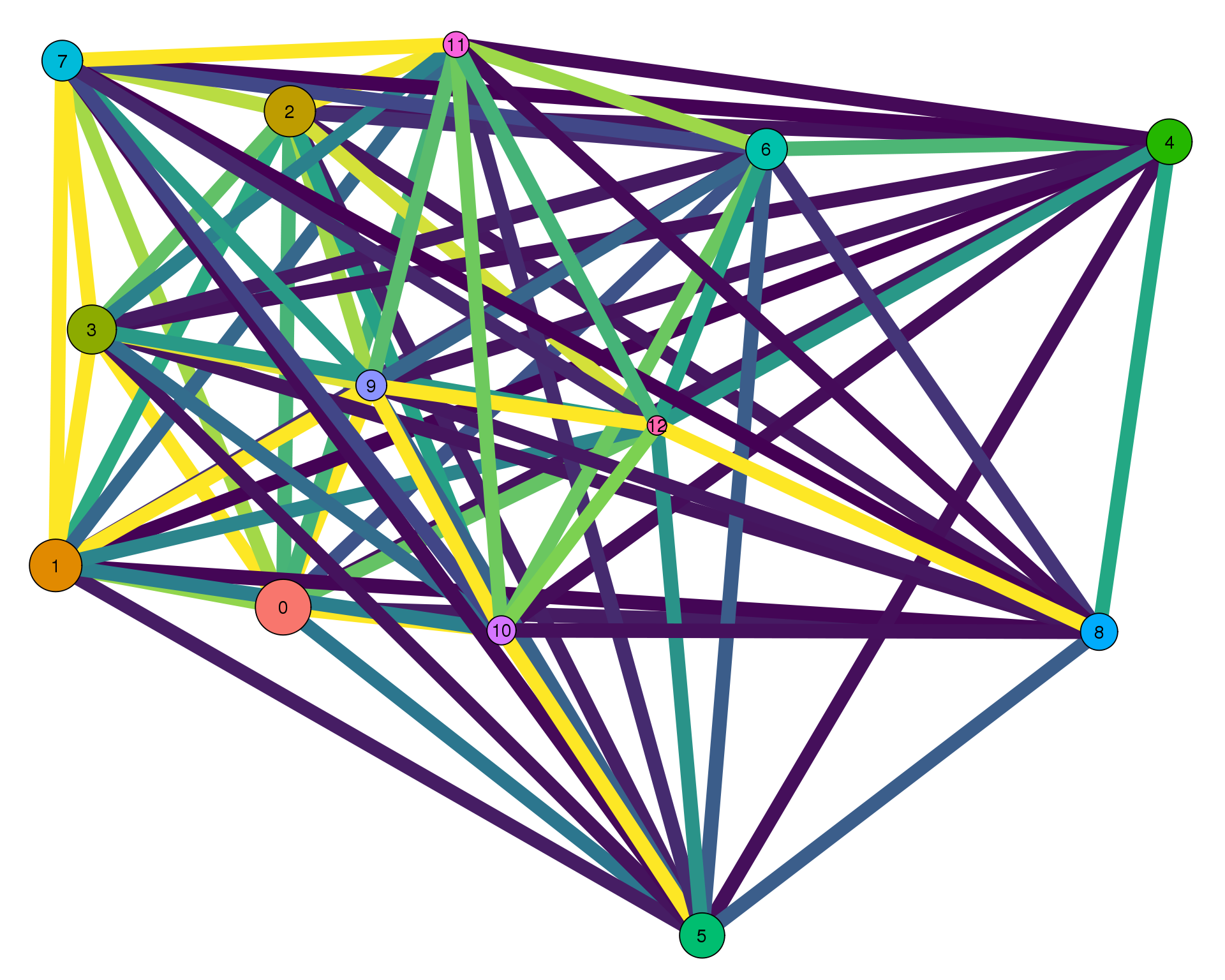
Expand here to see past versions of clust-paga-0-1.png:
| Version | Author | Date |
|---|---|---|
| fefdd07 | Luke Zappia | 2019-02-28 |
Con 0.1
plotPAGAClustGraph(clust_embedding, clust_edges, thresh = 0.1)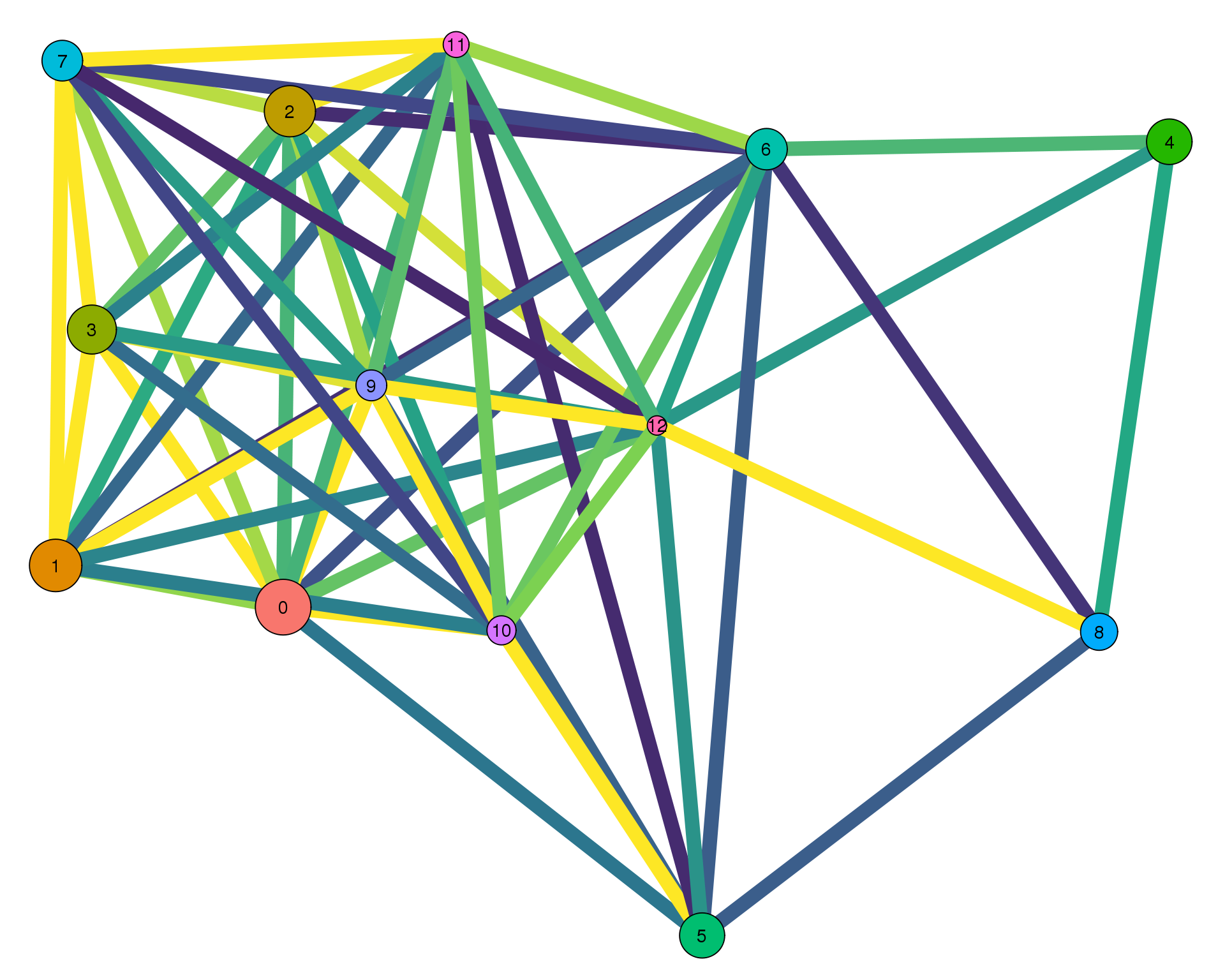
Expand here to see past versions of clust-paga-0.1-1.png:
| Version | Author | Date |
|---|---|---|
| fefdd07 | Luke Zappia | 2019-02-28 |
Con 0.2
plotPAGAClustGraph(clust_embedding, clust_edges, thresh = 0.2)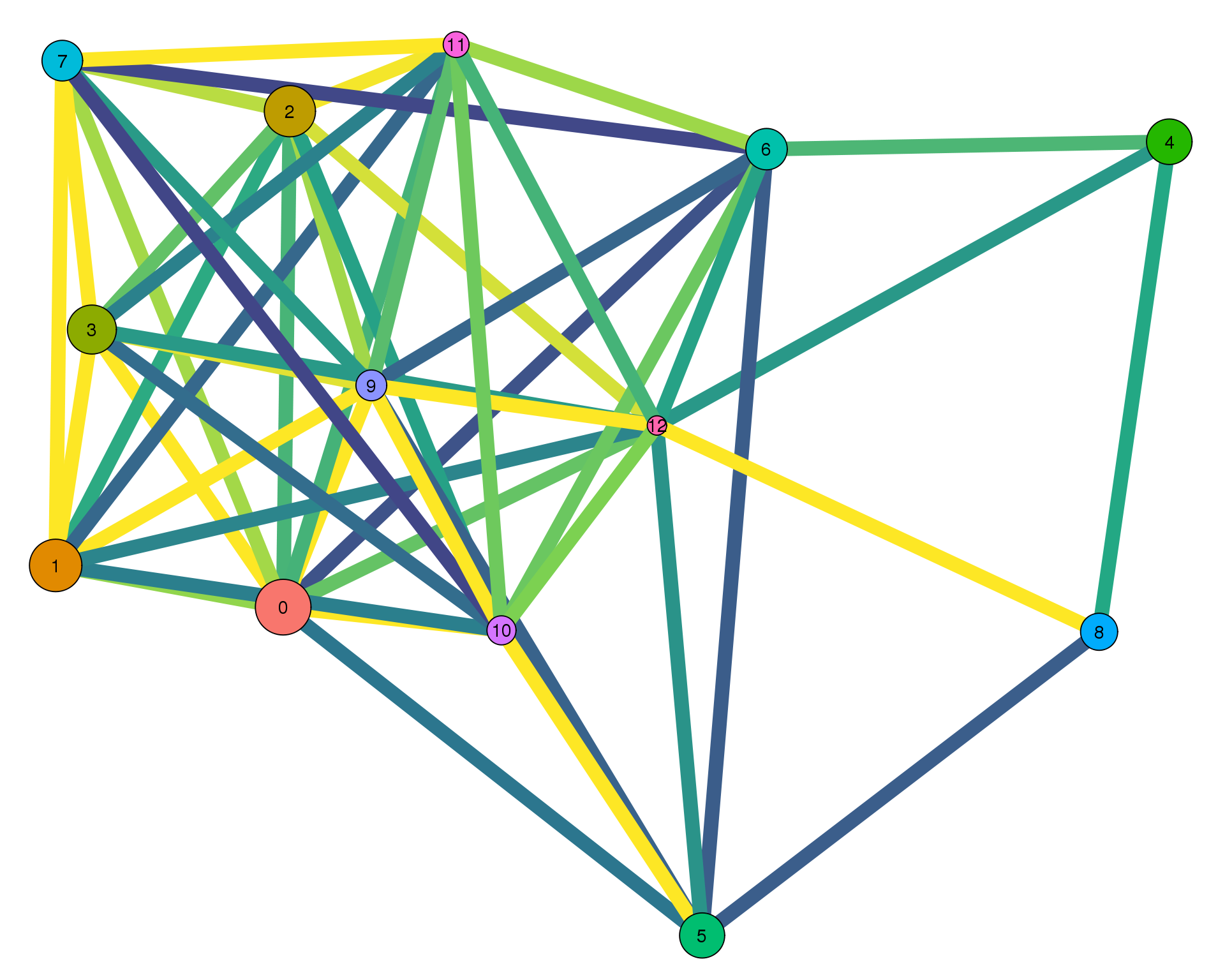
Expand here to see past versions of clust-paga-0.2-1.png:
| Version | Author | Date |
|---|---|---|
| fefdd07 | Luke Zappia | 2019-02-28 |
Con 0.3
plotPAGAClustGraph(clust_embedding, clust_edges, thresh = 0.3)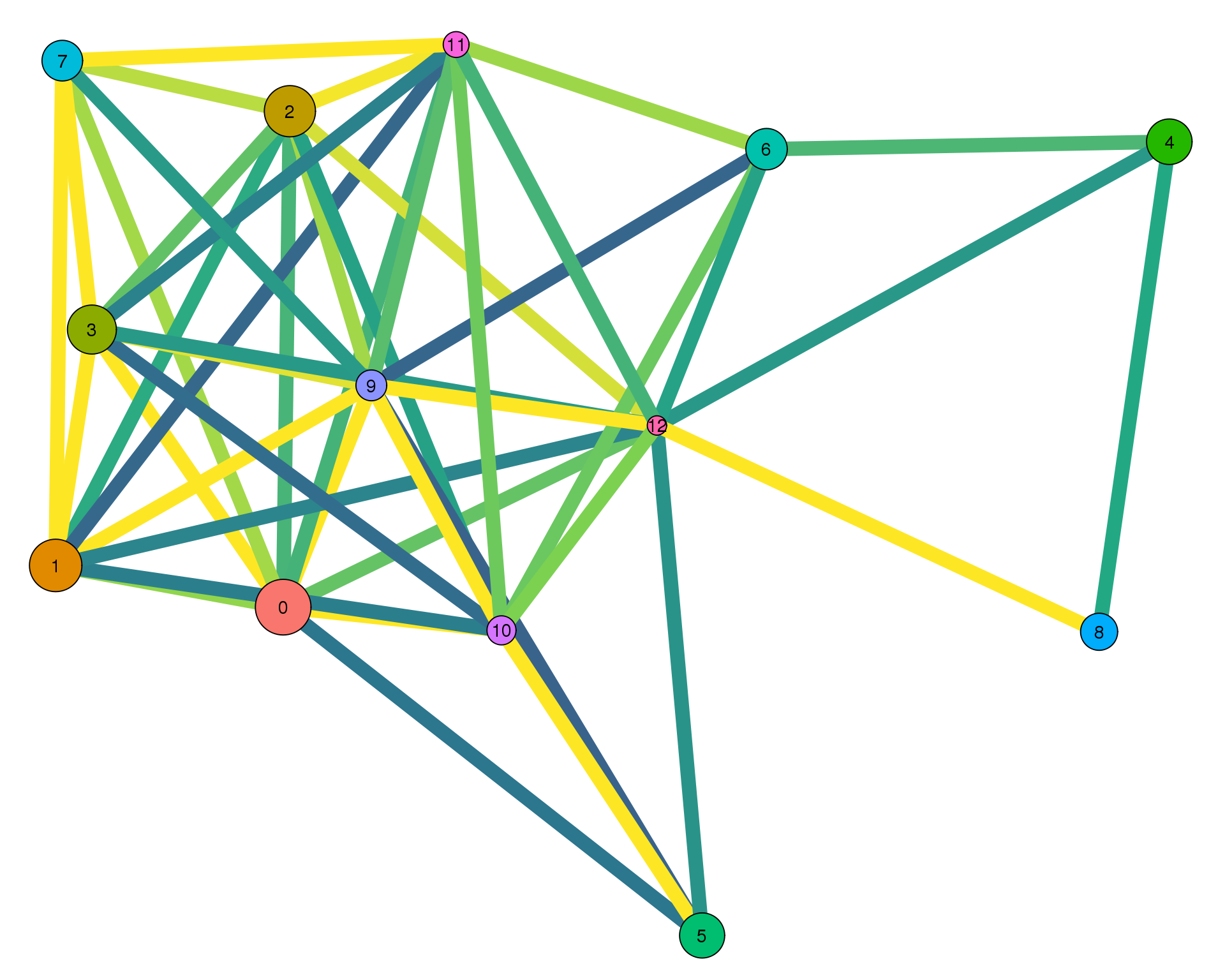
Expand here to see past versions of clust-paga-0.3-1.png:
| Version | Author | Date |
|---|---|---|
| fefdd07 | Luke Zappia | 2019-02-28 |
Con 0.4
plotPAGAClustGraph(clust_embedding, clust_edges, thresh = 0.4)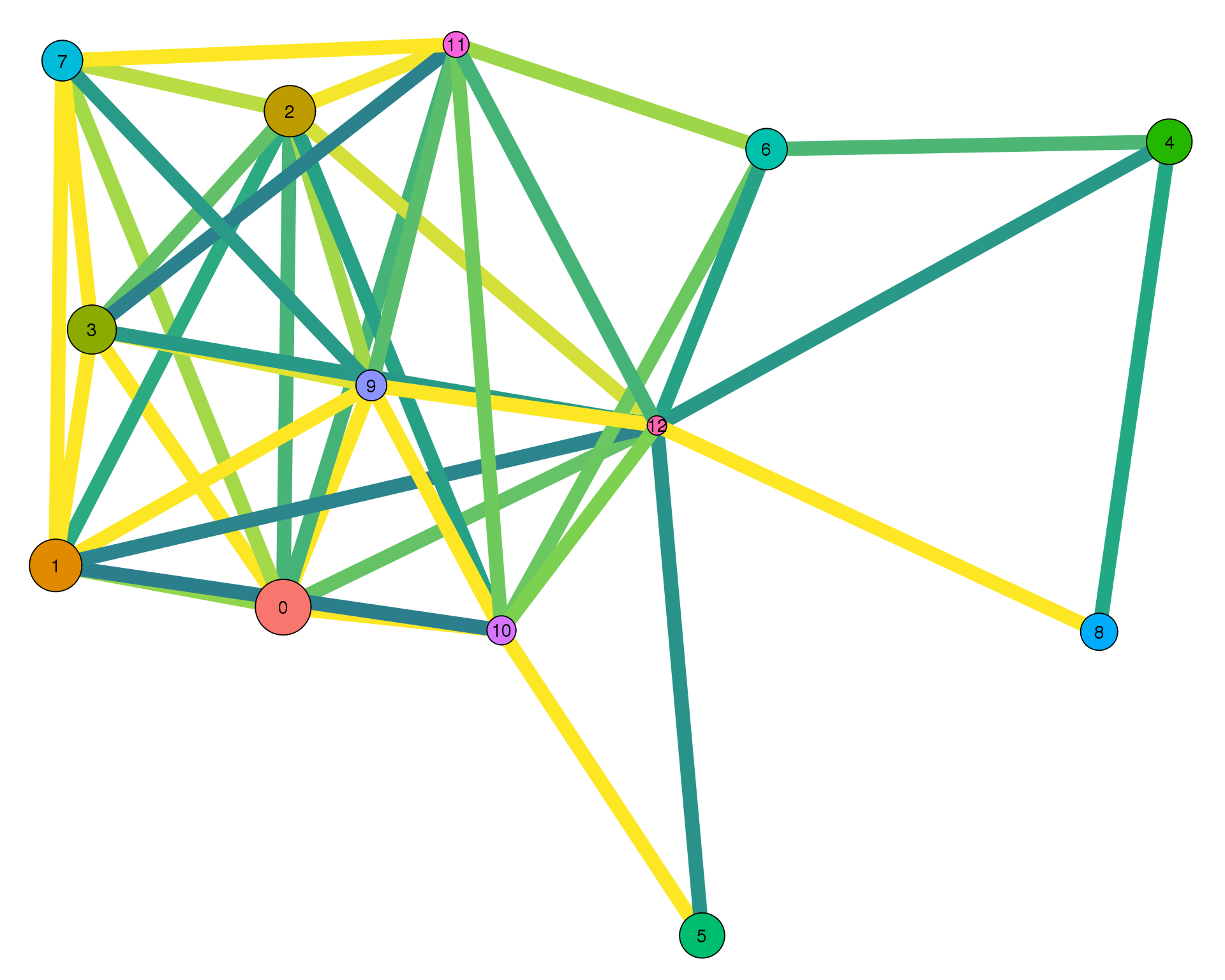
Expand here to see past versions of clust-paga-0.4-1.png:
| Version | Author | Date |
|---|---|---|
| fefdd07 | Luke Zappia | 2019-02-28 |
Con 0.5
plotPAGAClustGraph(clust_embedding, clust_edges, thresh = 0.5)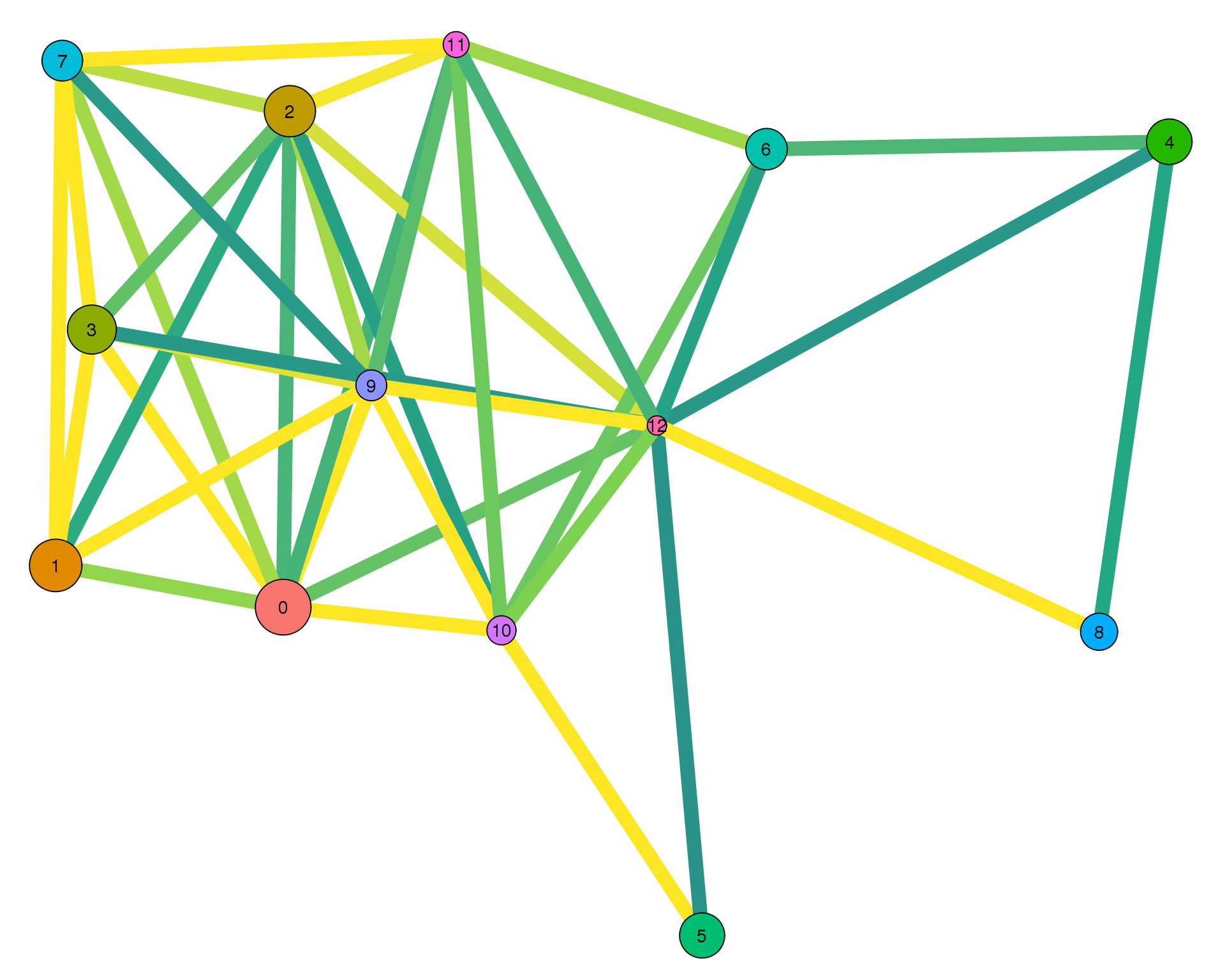
Expand here to see past versions of clust-paga-0.5-1.png:
| Version | Author | Date |
|---|---|---|
| fefdd07 | Luke Zappia | 2019-02-28 |
Con 0.6
plotPAGAClustGraph(clust_embedding, clust_edges, thresh = 0.6)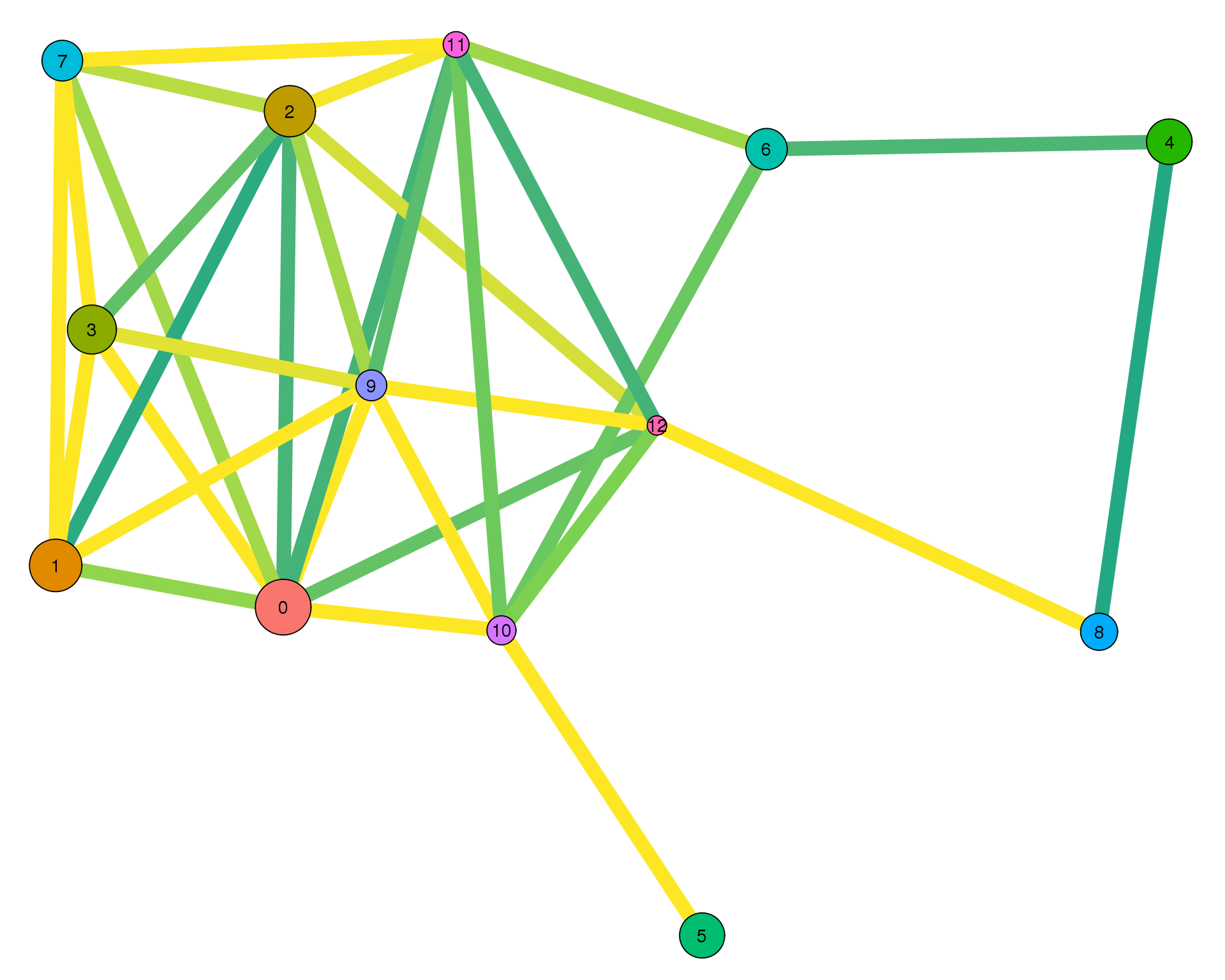
Expand here to see past versions of clust-paga-0.6-1.png:
| Version | Author | Date |
|---|---|---|
| fefdd07 | Luke Zappia | 2019-02-28 |
Con 0.7
plotPAGAClustGraph(clust_embedding, clust_edges, thresh = 0.7)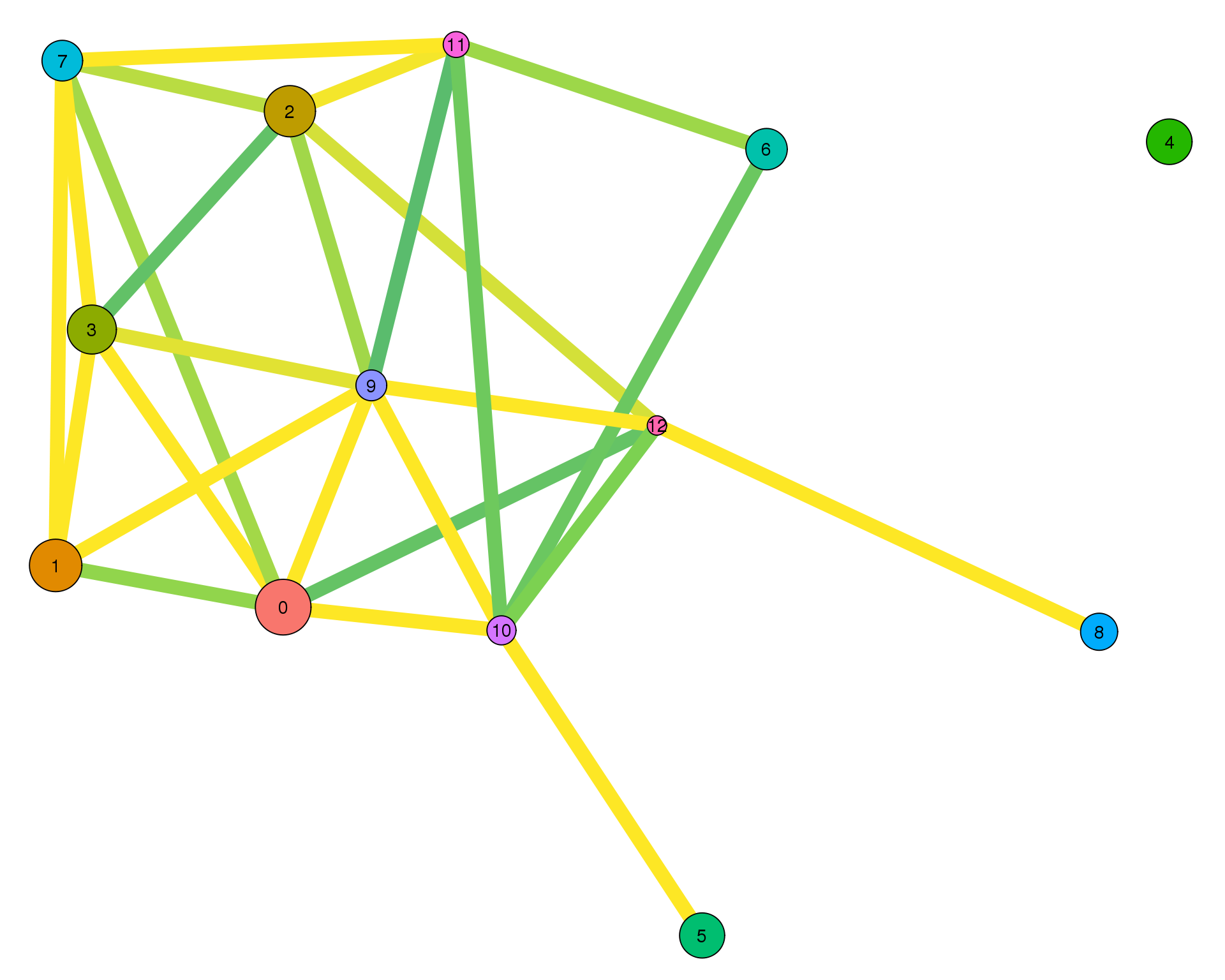
Expand here to see past versions of clust-paga-0.7-1.png:
| Version | Author | Date |
|---|---|---|
| fefdd07 | Luke Zappia | 2019-02-28 |
Con 0.8
plotPAGAClustGraph(clust_embedding, clust_edges, thresh = 0.8)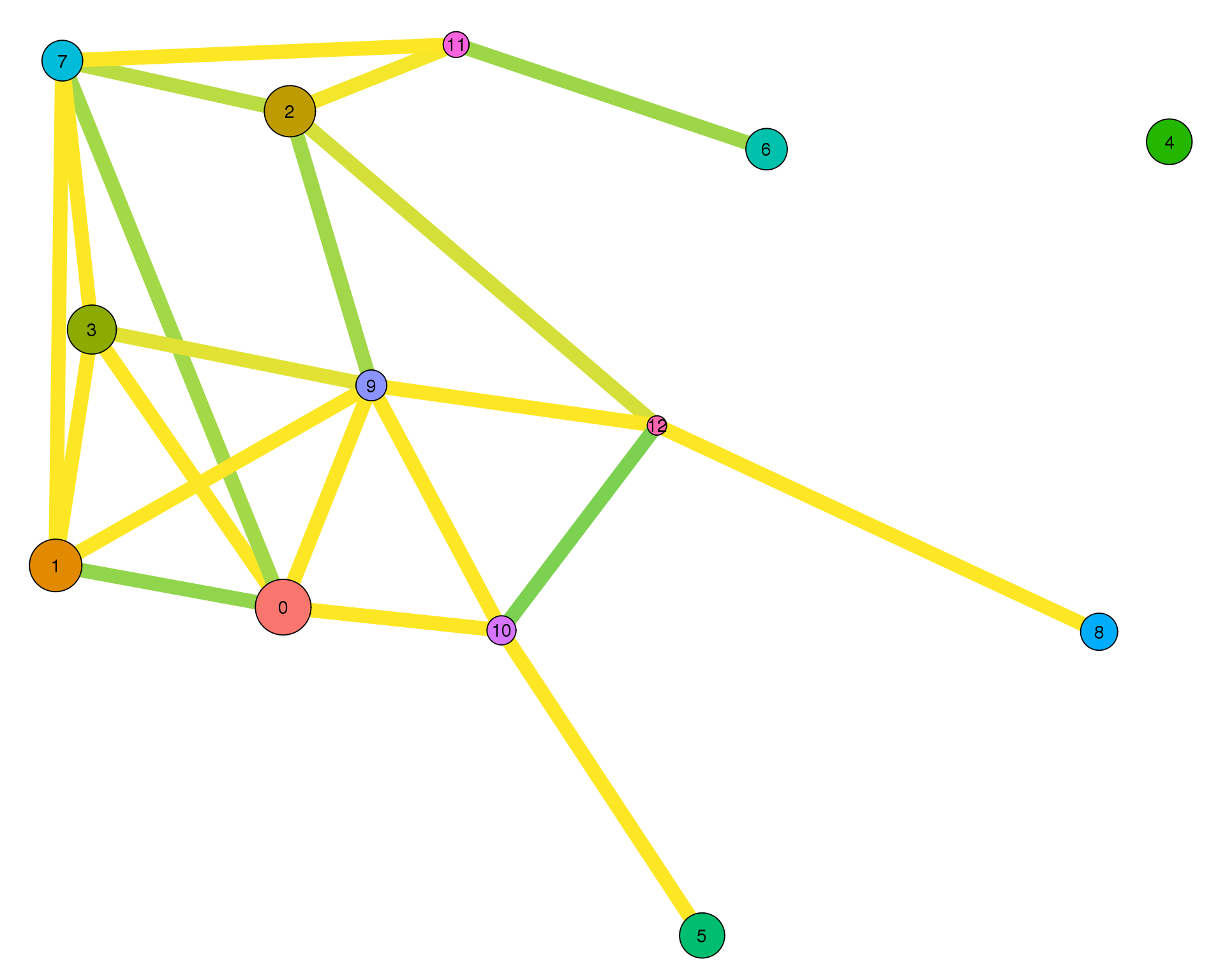
Expand here to see past versions of clust-paga-0.8-1.png:
| Version | Author | Date |
|---|---|---|
| fefdd07 | Luke Zappia | 2019-02-28 |
Con 0.9
plotPAGAClustGraph(clust_embedding, clust_edges, thresh = 0.9)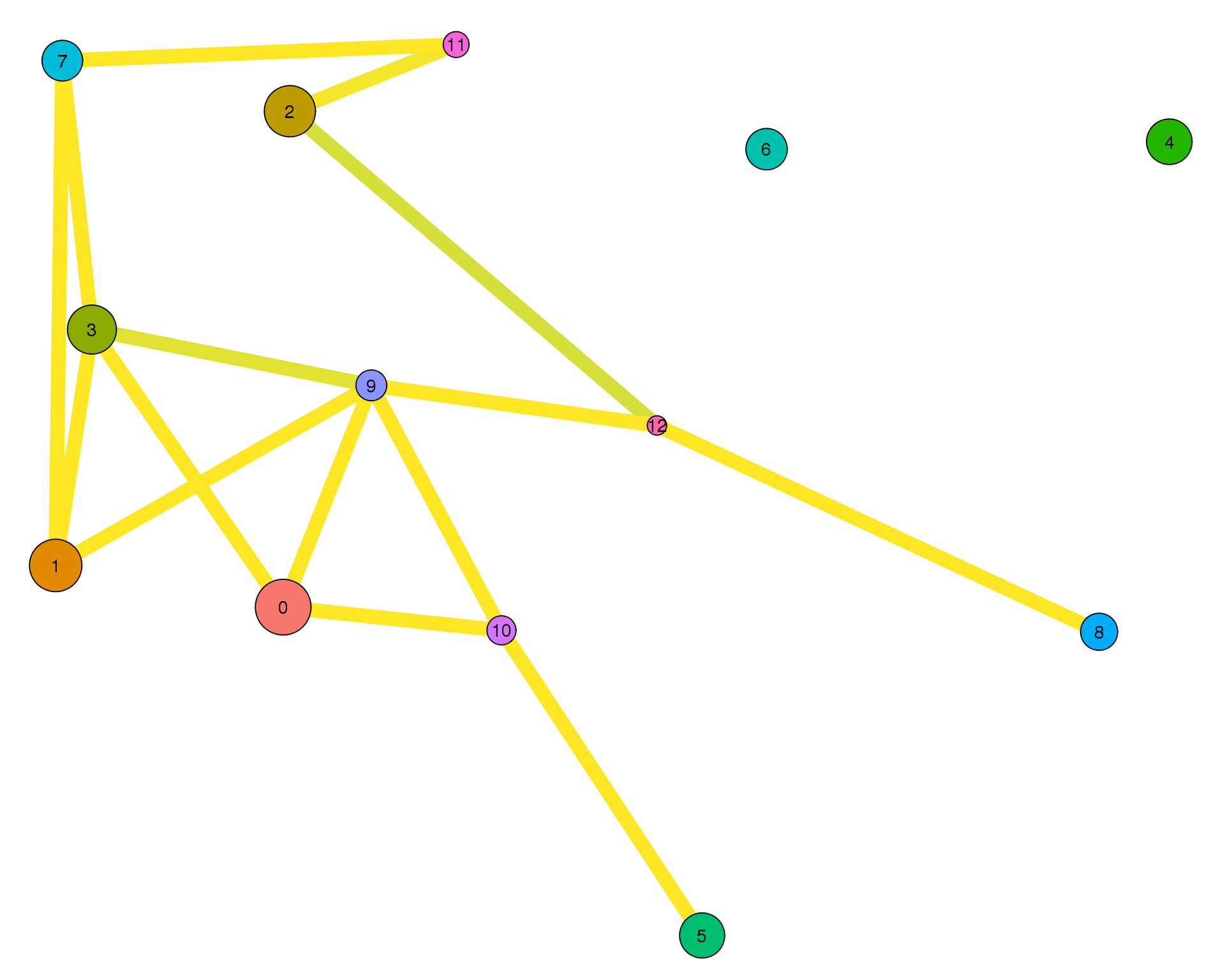
Expand here to see past versions of clust-paga-0.9-1.png:
| Version | Author | Date |
|---|---|---|
| fefdd07 | Luke Zappia | 2019-02-28 |
Edges by threshold
Number of selected edges for different threshold connectivities.
plot_data <- tibble(
Threshold = seq(0, 1, 0.01)
) %>%
mutate(Edges = map_int(Threshold, function(thresh) {
sum(clust_edges$Connectivity > thresh)
}))
con_thresh <- 0.7
ggplot(plot_data, aes(x = Threshold, y = Edges)) +
geom_point() +
geom_line() +
geom_vline(xintercept = con_thresh, colour = "red") +
xlab("Connectivity threshold") +
ylab("Number of edges") +
theme_minimal()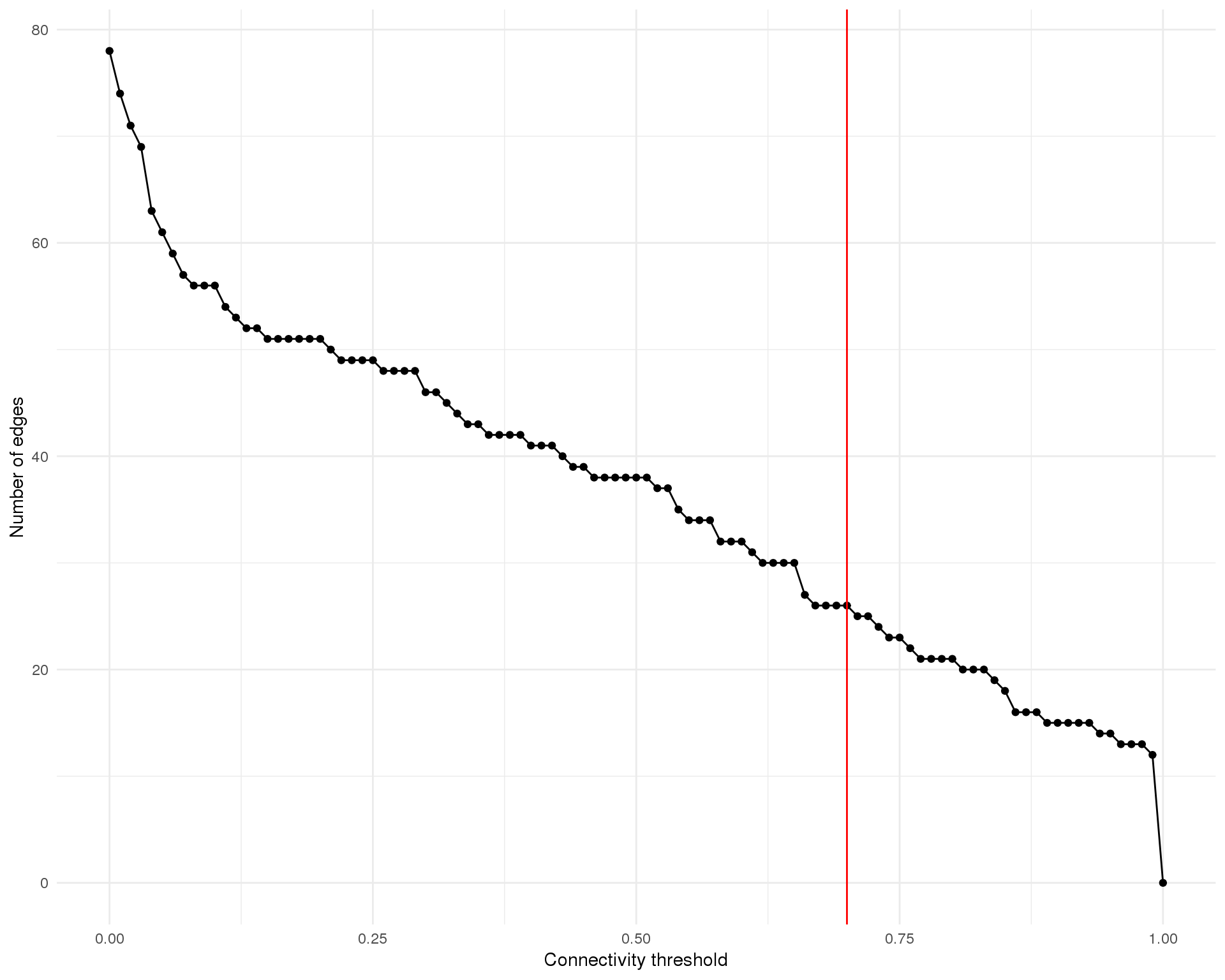
Expand here to see past versions of thresh-edges-1.png:
| Version | Author | Date |
|---|---|---|
| fefdd07 | Luke Zappia | 2019-02-28 |
Here we have selected a connectivity threshold of 0.7.
Cell graph
The cluster graph can be used as a starting point to layout individual cells, allowing us to see things at a higher resolution.
plotPAGACellGraph(cell_embedding, cell_edges, thresh = 0.1)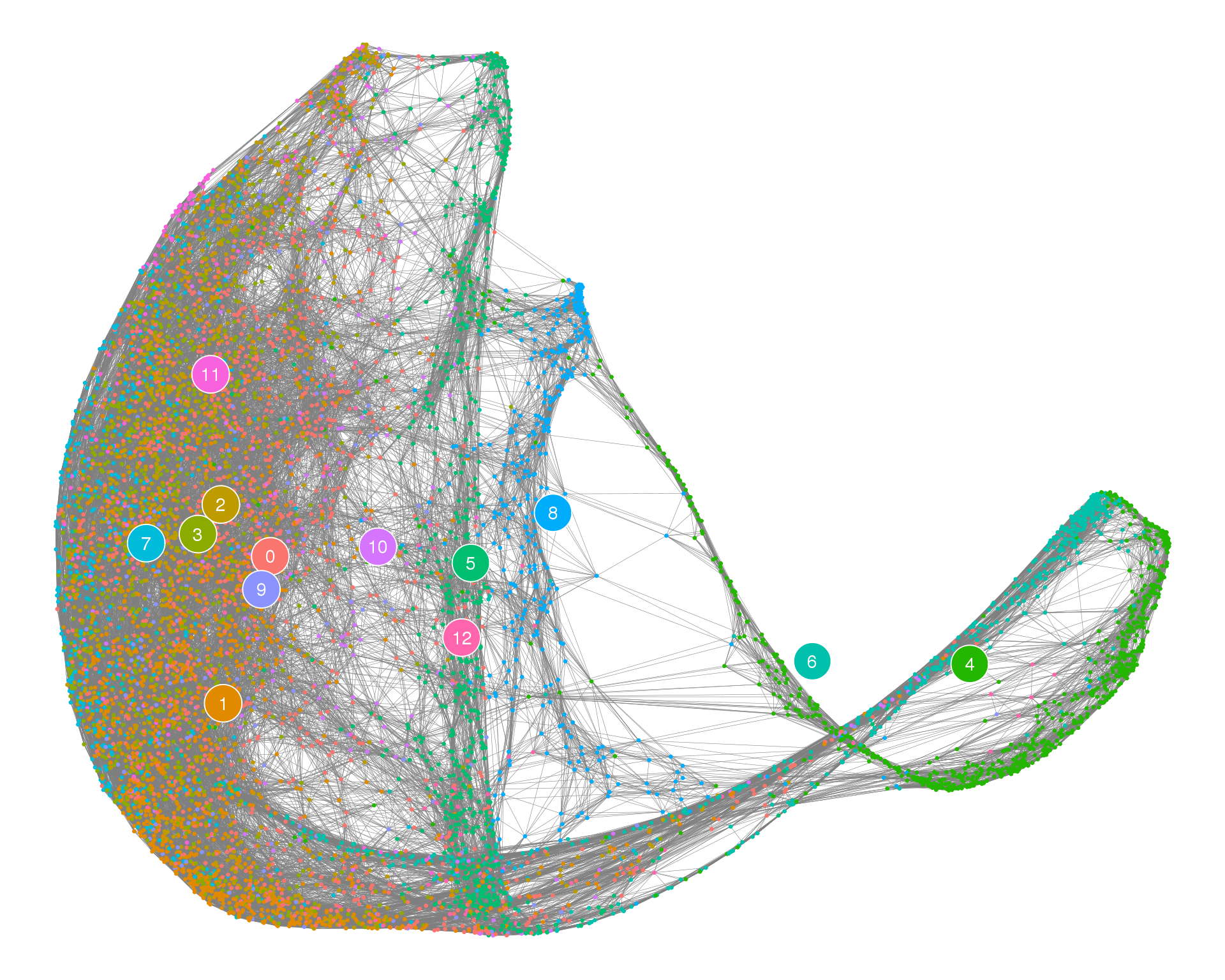
Expand here to see past versions of cell-paga-1.png:
| Version | Author | Date |
|---|---|---|
| fefdd07 | Luke Zappia | 2019-02-28 |
Compare
Looking the two views together let’s us see both global and specific details.
plotPAGACompare(clust_embedding, clust_edges, clust_thresh = con_thresh,
cell_embedding, cell_edges, cell_thresh = 0.1)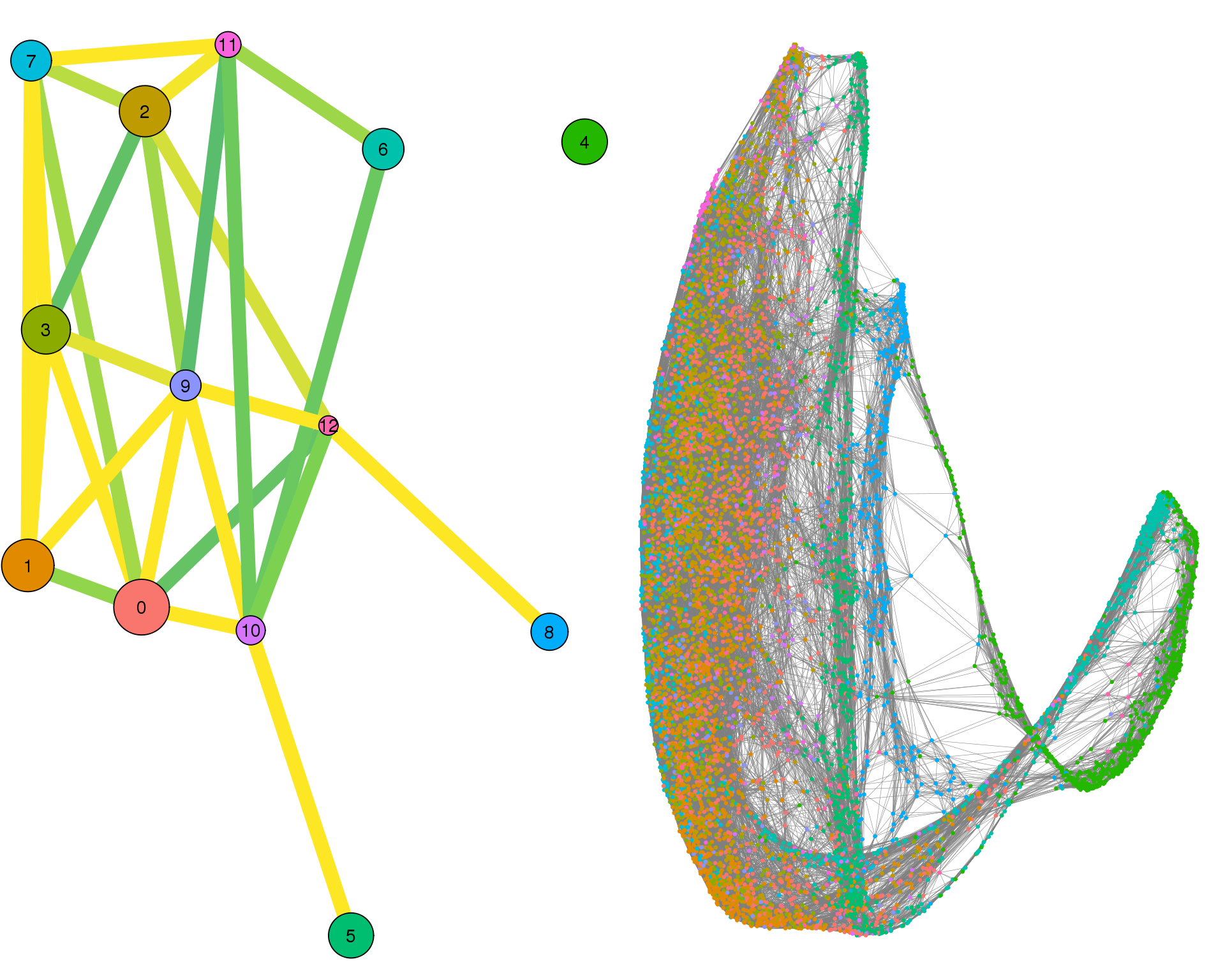
Expand here to see past versions of compare-paga-1.png:
| Version | Author | Date |
|---|---|---|
| fefdd07 | Luke Zappia | 2019-02-28 |
Genes
known_genes <- c(
# Stroma
"TAGLN", "ACTA2", "MAB21L2", "DLK1", "GATA3", "COL2A1", "COL9A3",
# Podocyte
"PODXL", "NPHS2", "TCF21",
# Cell cycle
"HIST1H4C", "PCLAF", "CENPF", "HMGB2",
# Endothelium
"CLDN5", "PECAM1", "KDR", "CALM1",
# Neural
"TTYH1", "SOX2", "HES6", "STMN2",
# Epithelium
"PAX2", "PAX8", "KRT19",
# Muscle
"MYOG", "MYOD1"
)
for (gene in known_genes) {
cell_embedding[[gene]] <- logcounts(sce)[gene, ]
}
clust_genes <- cell_embedding %>%
select(-Cell, -X, -Y) %>%
group_by(Cluster) %>%
summarise_all(mean)
clust_embedding <- left_join(clust_embedding, clust_genes, by = "Cluster")
src_list <- lapply(known_genes, function(gene) {
src <- c(
"### {{gene}} {.unnumbered}",
"```{r compare-{{gene}}}",
"plotPAGACompare(clust_embedding, clust_edges,",
"clust_thresh = con_thresh, cell_embedding, cell_edges,
cell_thresh = 0.1, colour = '{{gene}}')",
"```",
""
)
knit_expand(text = src)
})
out <- knit_child(text = unlist(src_list), options = list(cache = FALSE))TAGLN
plotPAGACompare(clust_embedding, clust_edges,
clust_thresh = con_thresh, cell_embedding, cell_edges,
cell_thresh = 0.1, colour = 'TAGLN')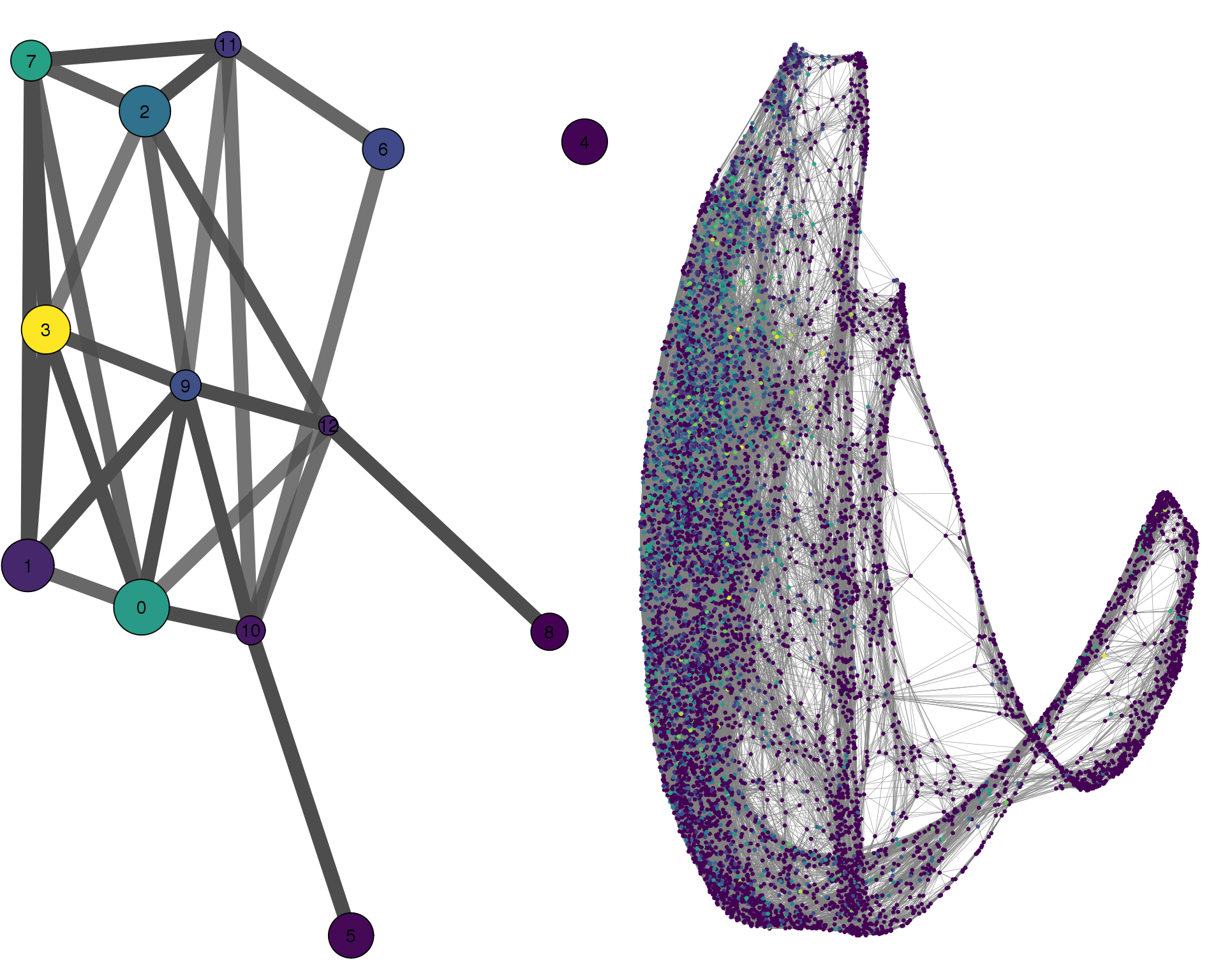
Expand here to see past versions of compare-TAGLN-1.png:
| Version | Author | Date |
|---|---|---|
| fefdd07 | Luke Zappia | 2019-02-28 |
ACTA2
plotPAGACompare(clust_embedding, clust_edges,
clust_thresh = con_thresh, cell_embedding, cell_edges,
cell_thresh = 0.1, colour = 'ACTA2')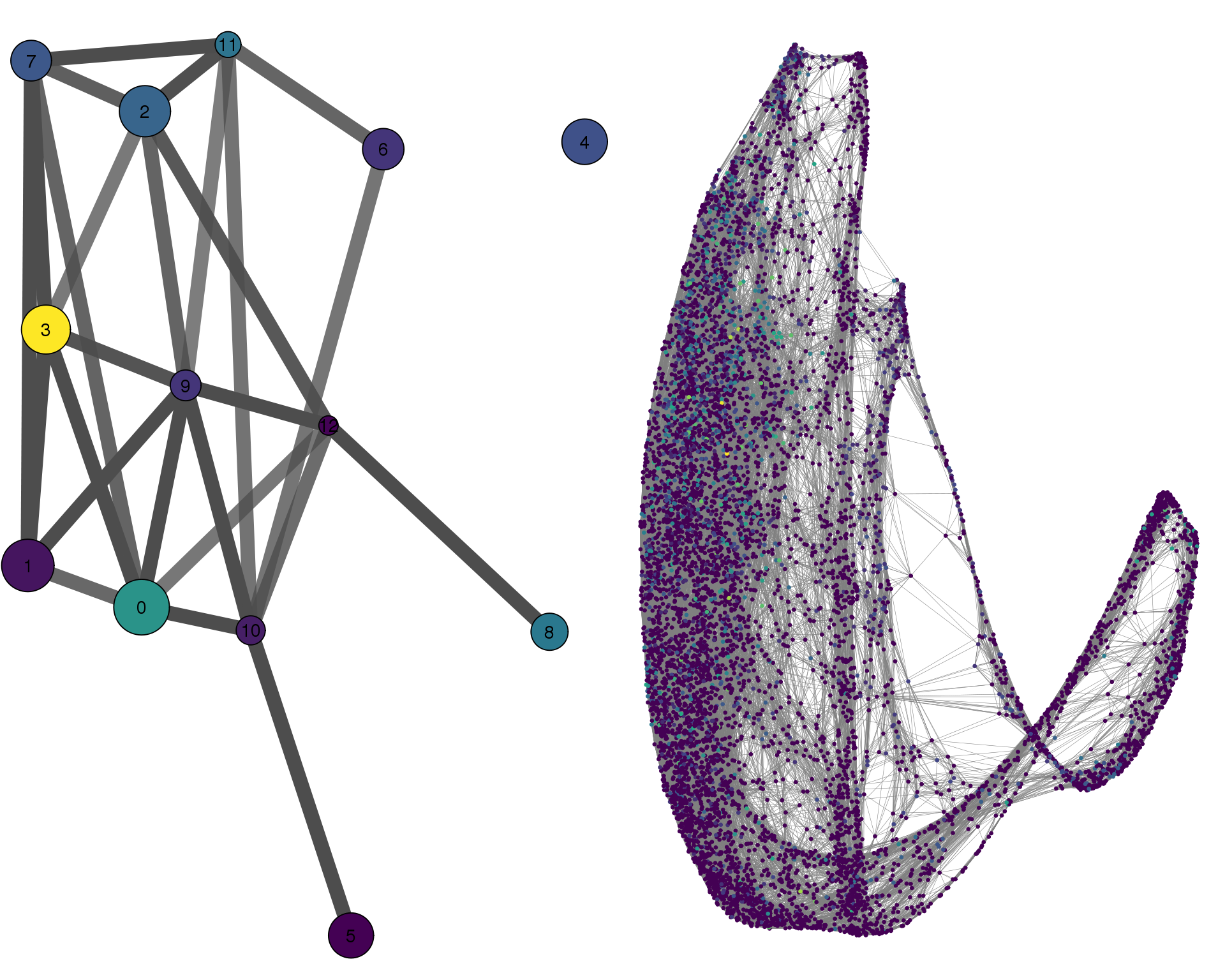
Expand here to see past versions of compare-ACTA2-1.png:
| Version | Author | Date |
|---|---|---|
| fefdd07 | Luke Zappia | 2019-02-28 |
MAB21L2
plotPAGACompare(clust_embedding, clust_edges,
clust_thresh = con_thresh, cell_embedding, cell_edges,
cell_thresh = 0.1, colour = 'MAB21L2')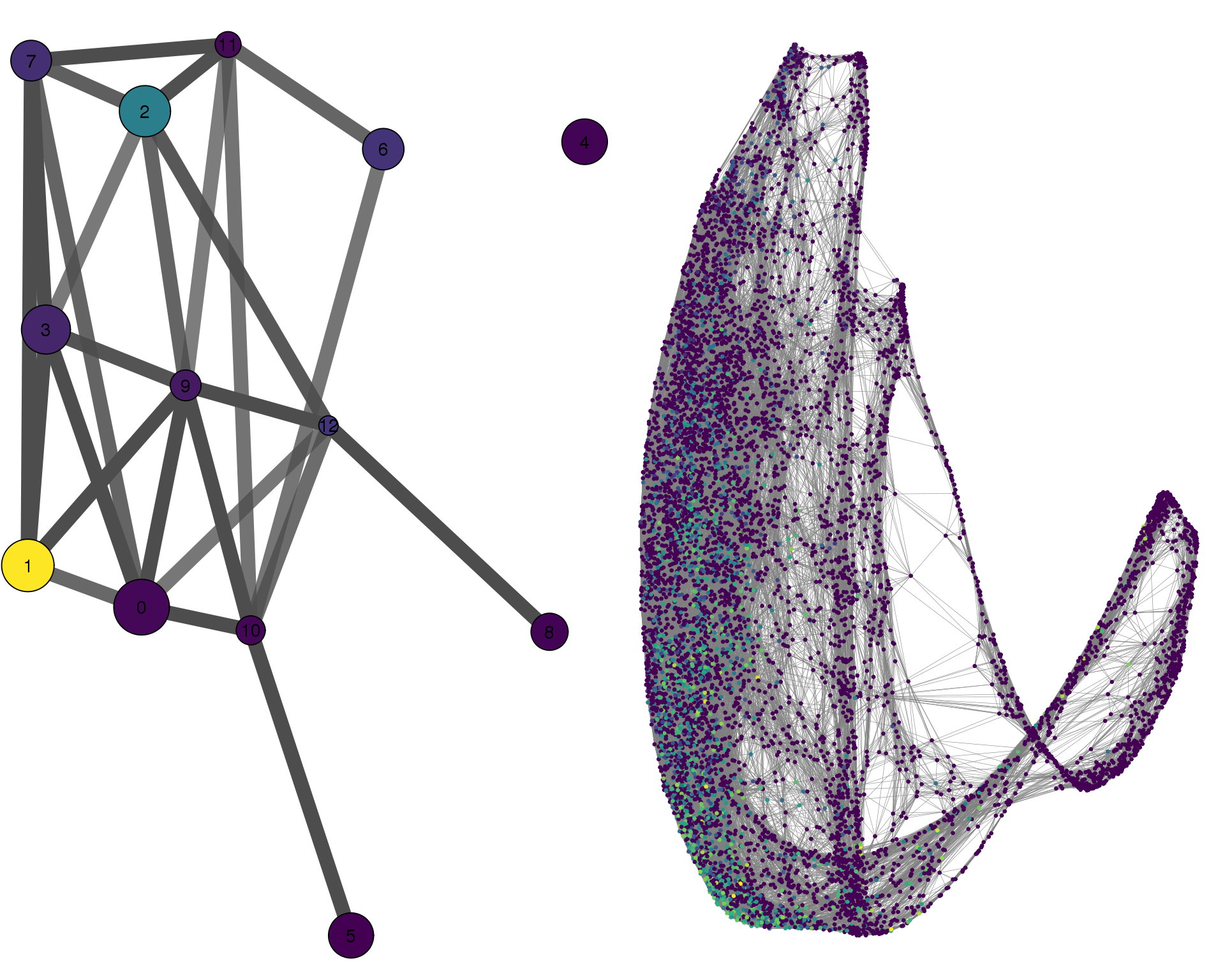
Expand here to see past versions of compare-MAB21L2-1.png:
| Version | Author | Date |
|---|---|---|
| fefdd07 | Luke Zappia | 2019-02-28 |
DLK1
plotPAGACompare(clust_embedding, clust_edges,
clust_thresh = con_thresh, cell_embedding, cell_edges,
cell_thresh = 0.1, colour = 'DLK1')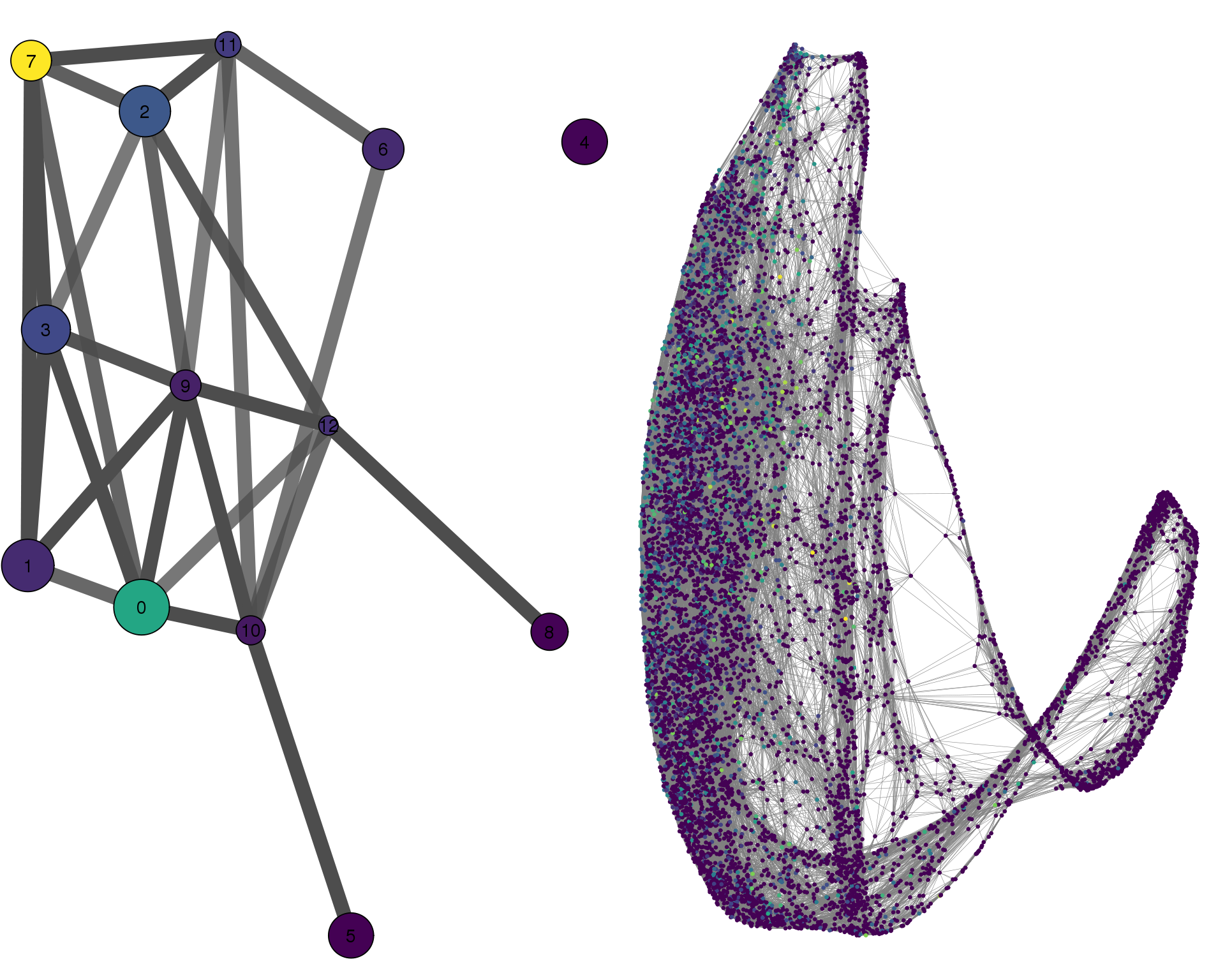
Expand here to see past versions of compare-DLK1-1.png:
| Version | Author | Date |
|---|---|---|
| fefdd07 | Luke Zappia | 2019-02-28 |
GATA3
plotPAGACompare(clust_embedding, clust_edges,
clust_thresh = con_thresh, cell_embedding, cell_edges,
cell_thresh = 0.1, colour = 'GATA3')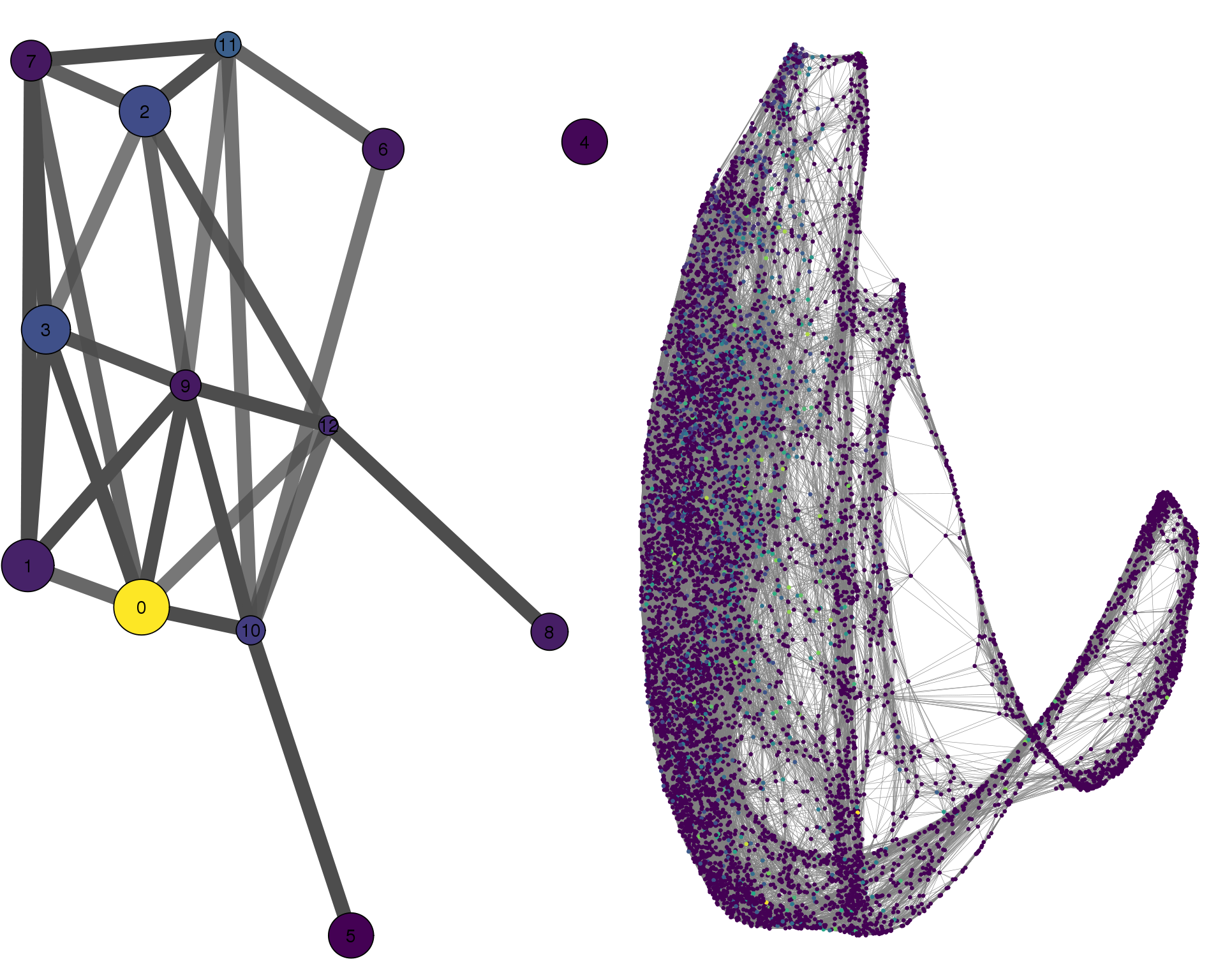
Expand here to see past versions of compare-GATA3-1.png:
| Version | Author | Date |
|---|---|---|
| fefdd07 | Luke Zappia | 2019-02-28 |
COL2A1
plotPAGACompare(clust_embedding, clust_edges,
clust_thresh = con_thresh, cell_embedding, cell_edges,
cell_thresh = 0.1, colour = 'COL2A1')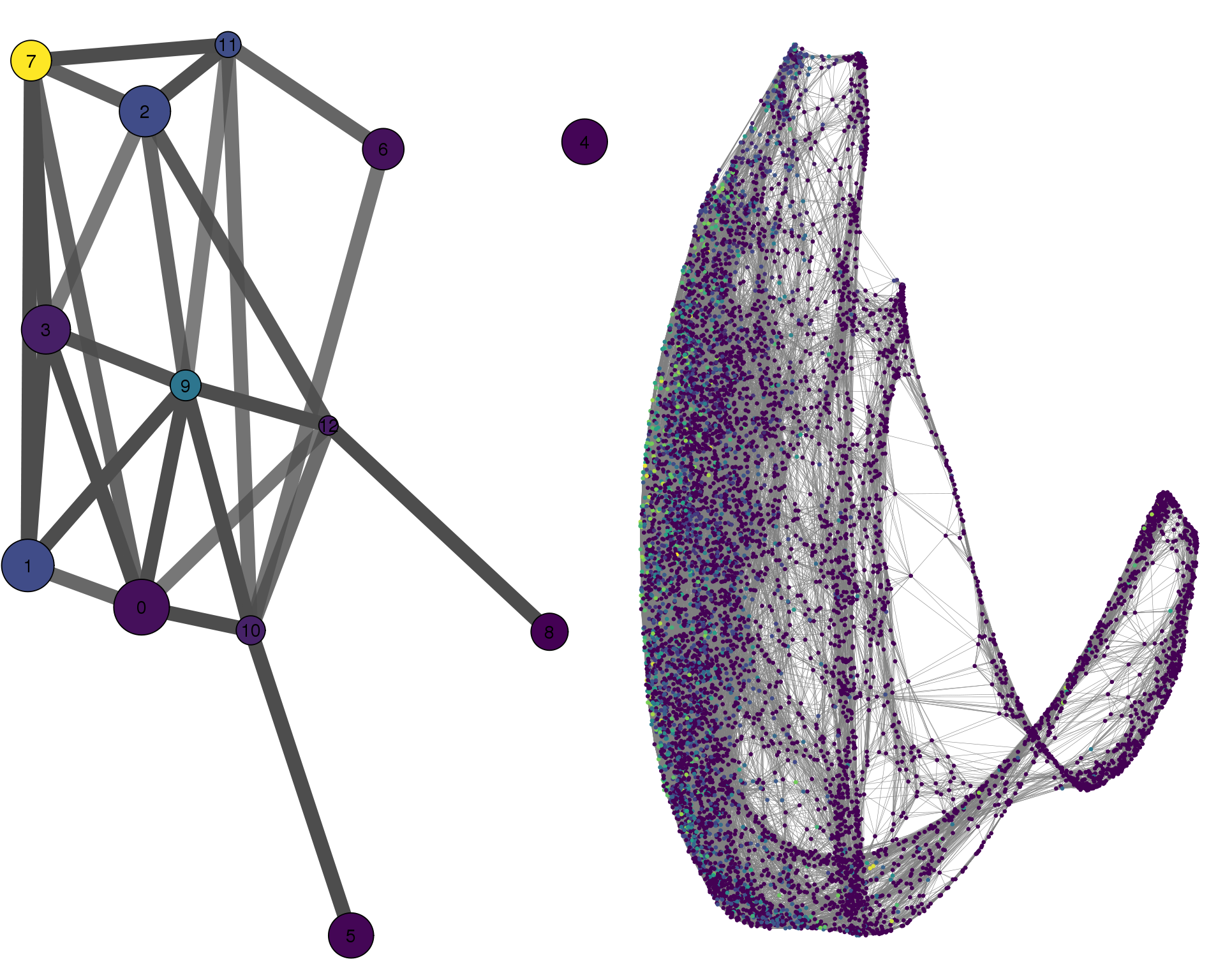
Expand here to see past versions of compare-COL2A1-1.png:
| Version | Author | Date |
|---|---|---|
| fefdd07 | Luke Zappia | 2019-02-28 |
COL9A3
plotPAGACompare(clust_embedding, clust_edges,
clust_thresh = con_thresh, cell_embedding, cell_edges,
cell_thresh = 0.1, colour = 'COL9A3')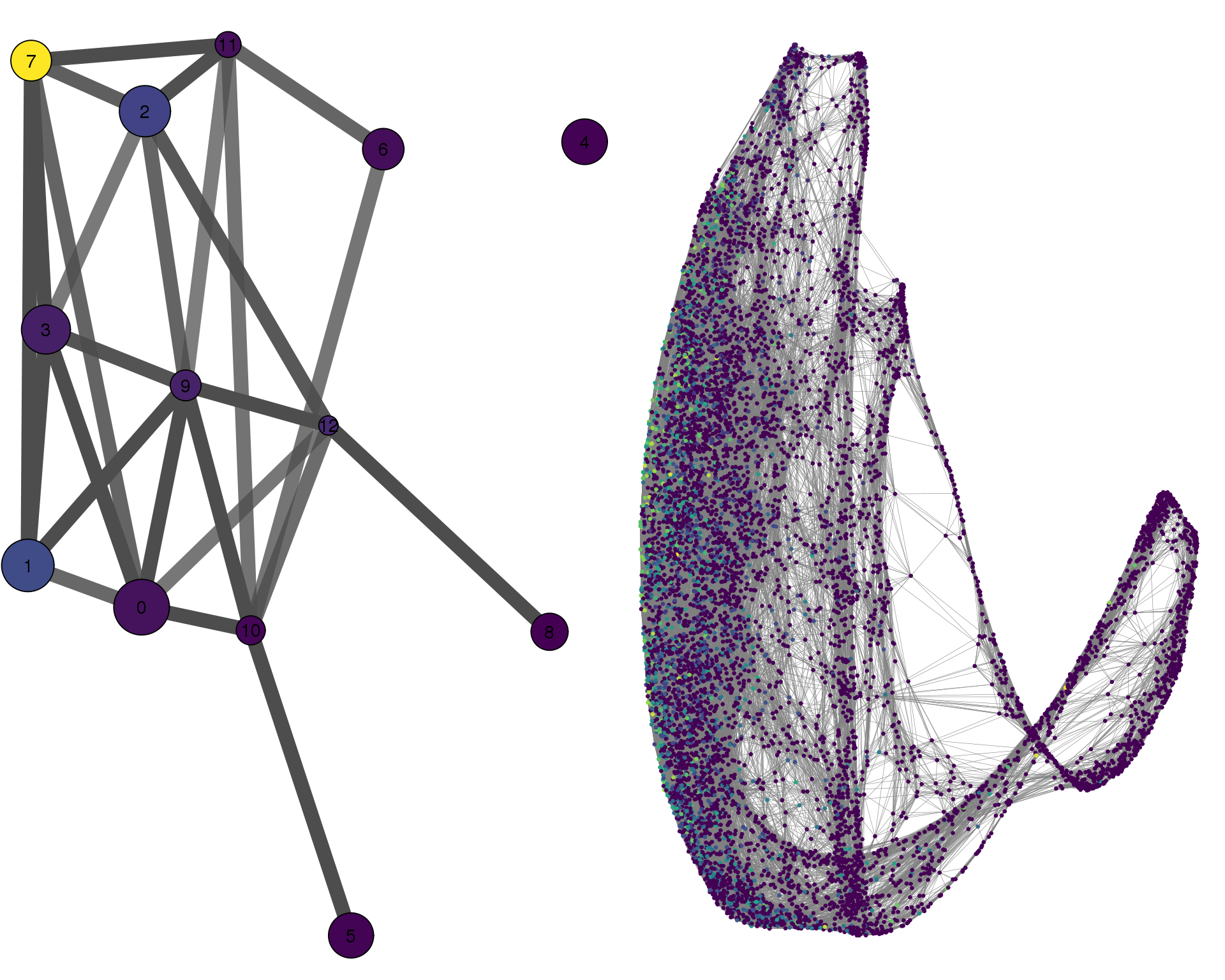
Expand here to see past versions of compare-COL9A3-1.png:
| Version | Author | Date |
|---|---|---|
| fefdd07 | Luke Zappia | 2019-02-28 |
PODXL
plotPAGACompare(clust_embedding, clust_edges,
clust_thresh = con_thresh, cell_embedding, cell_edges,
cell_thresh = 0.1, colour = 'PODXL')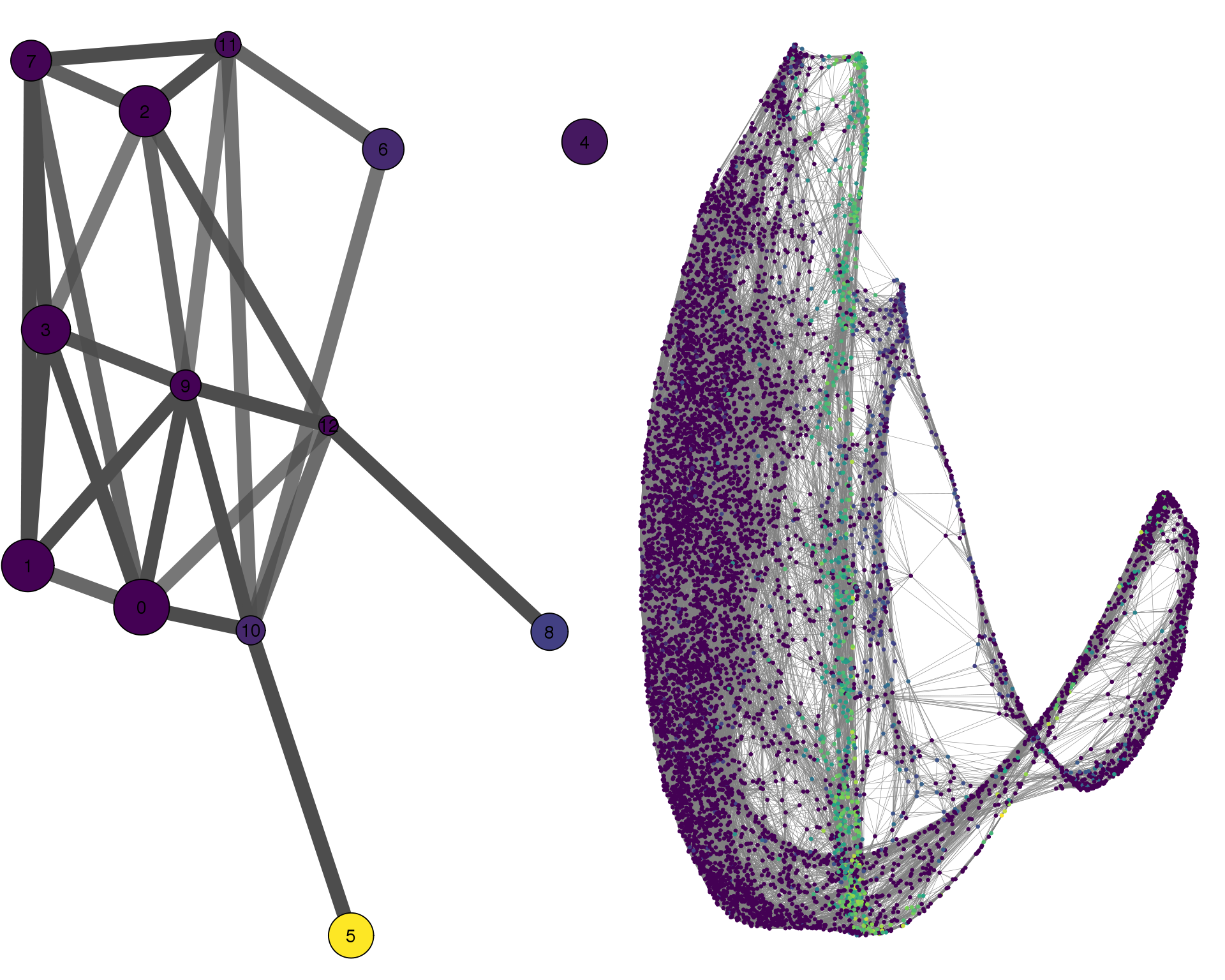
Expand here to see past versions of compare-PODXL-1.png:
| Version | Author | Date |
|---|---|---|
| fefdd07 | Luke Zappia | 2019-02-28 |
NPHS2
plotPAGACompare(clust_embedding, clust_edges,
clust_thresh = con_thresh, cell_embedding, cell_edges,
cell_thresh = 0.1, colour = 'NPHS2')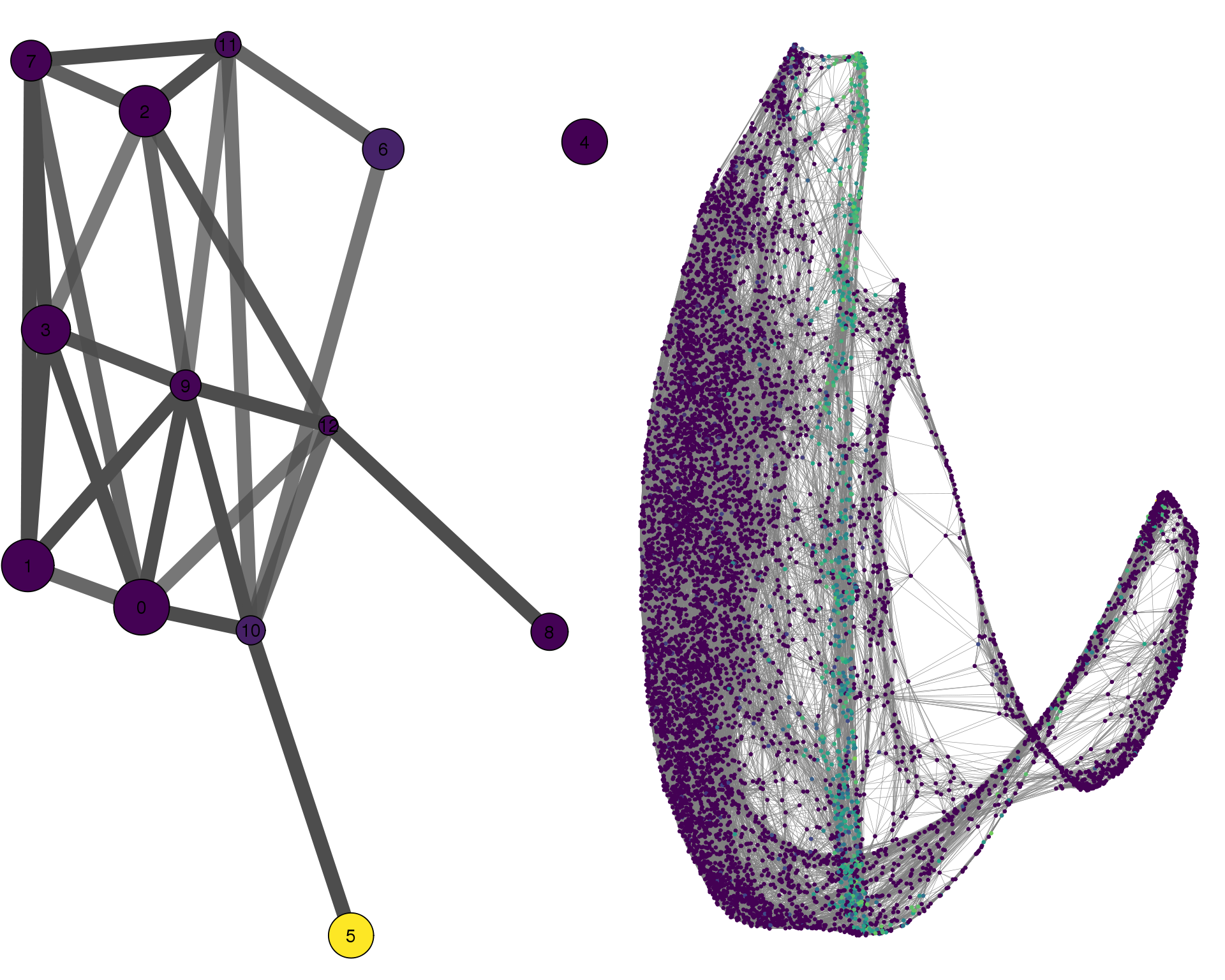
Expand here to see past versions of compare-NPHS2-1.png:
| Version | Author | Date |
|---|---|---|
| fefdd07 | Luke Zappia | 2019-02-28 |
TCF21
plotPAGACompare(clust_embedding, clust_edges,
clust_thresh = con_thresh, cell_embedding, cell_edges,
cell_thresh = 0.1, colour = 'TCF21')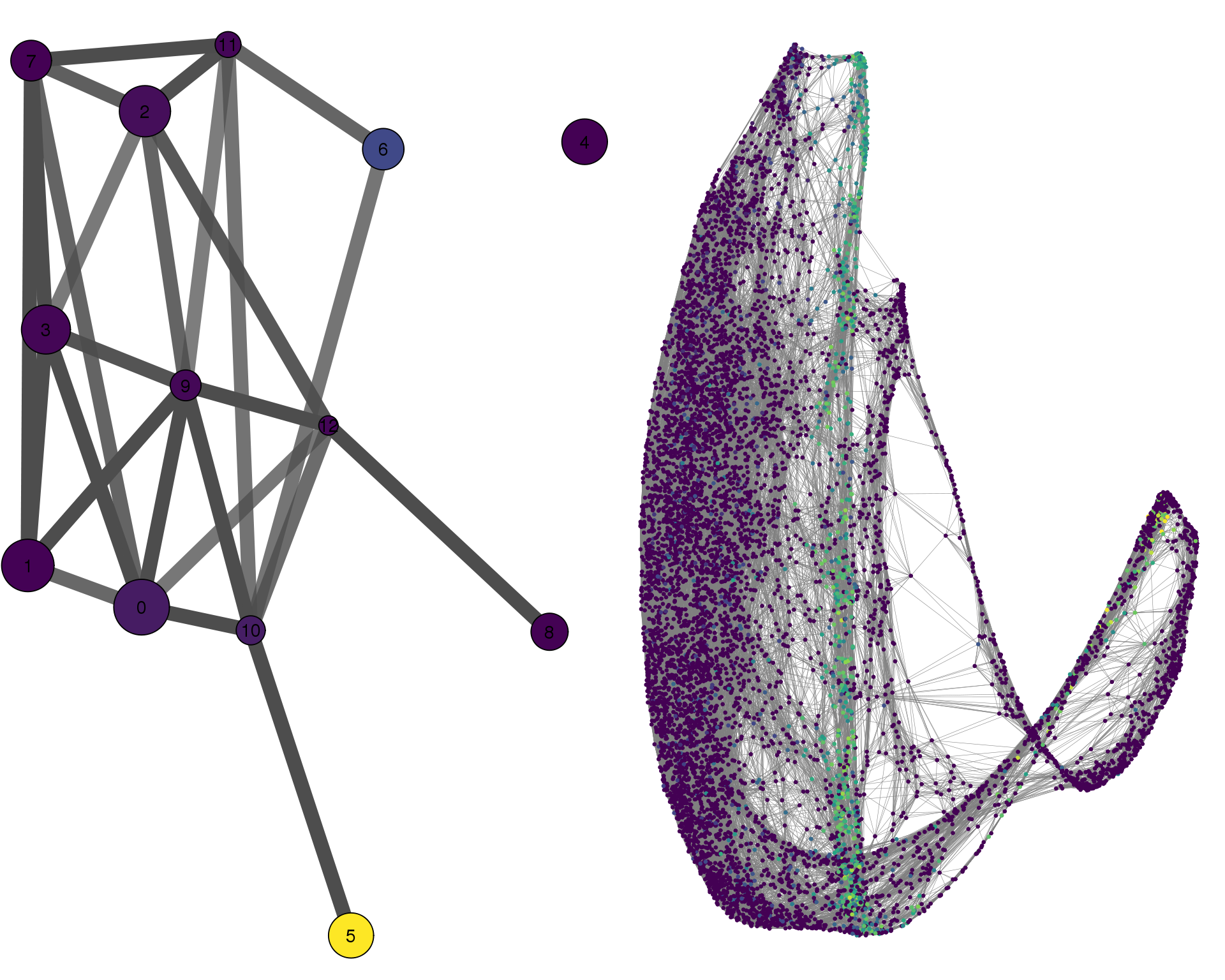
Expand here to see past versions of compare-TCF21-1.png:
| Version | Author | Date |
|---|---|---|
| fefdd07 | Luke Zappia | 2019-02-28 |
HIST1H4C
plotPAGACompare(clust_embedding, clust_edges,
clust_thresh = con_thresh, cell_embedding, cell_edges,
cell_thresh = 0.1, colour = 'HIST1H4C')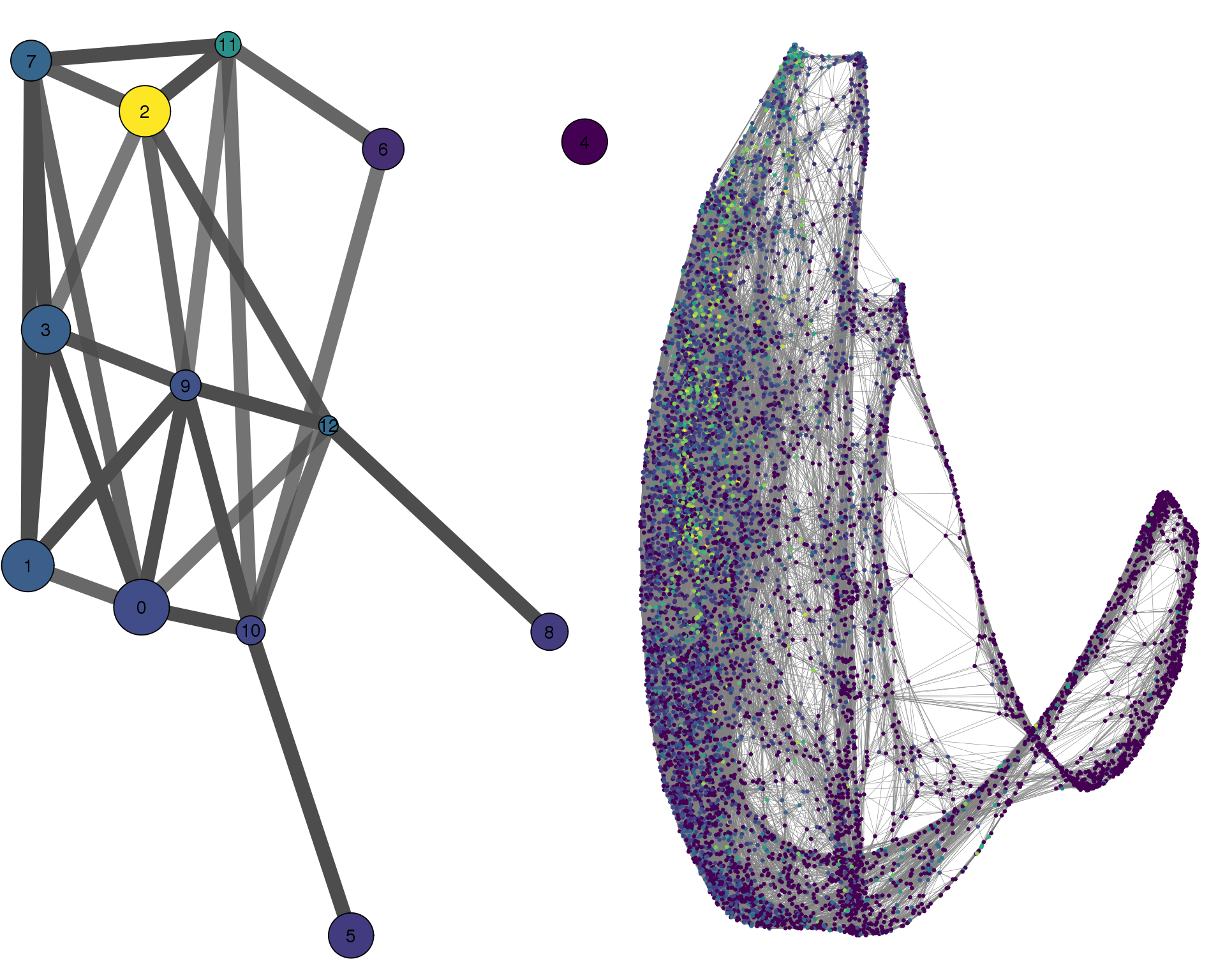
Expand here to see past versions of compare-HIST1H4C-1.png:
| Version | Author | Date |
|---|---|---|
| fefdd07 | Luke Zappia | 2019-02-28 |
PCLAF
plotPAGACompare(clust_embedding, clust_edges,
clust_thresh = con_thresh, cell_embedding, cell_edges,
cell_thresh = 0.1, colour = 'PCLAF')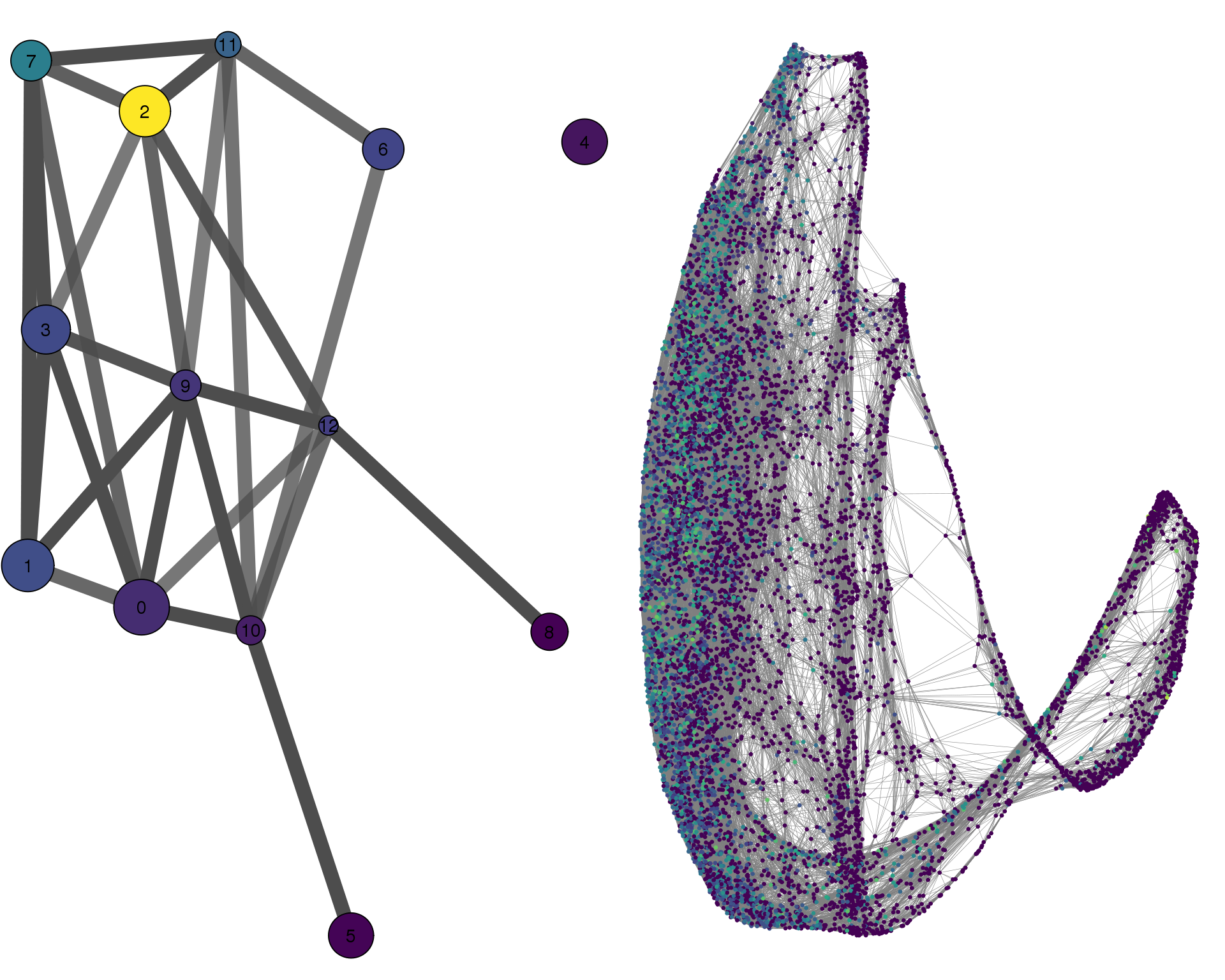
Expand here to see past versions of compare-PCLAF-1.png:
| Version | Author | Date |
|---|---|---|
| fefdd07 | Luke Zappia | 2019-02-28 |
CENPF
plotPAGACompare(clust_embedding, clust_edges,
clust_thresh = con_thresh, cell_embedding, cell_edges,
cell_thresh = 0.1, colour = 'CENPF')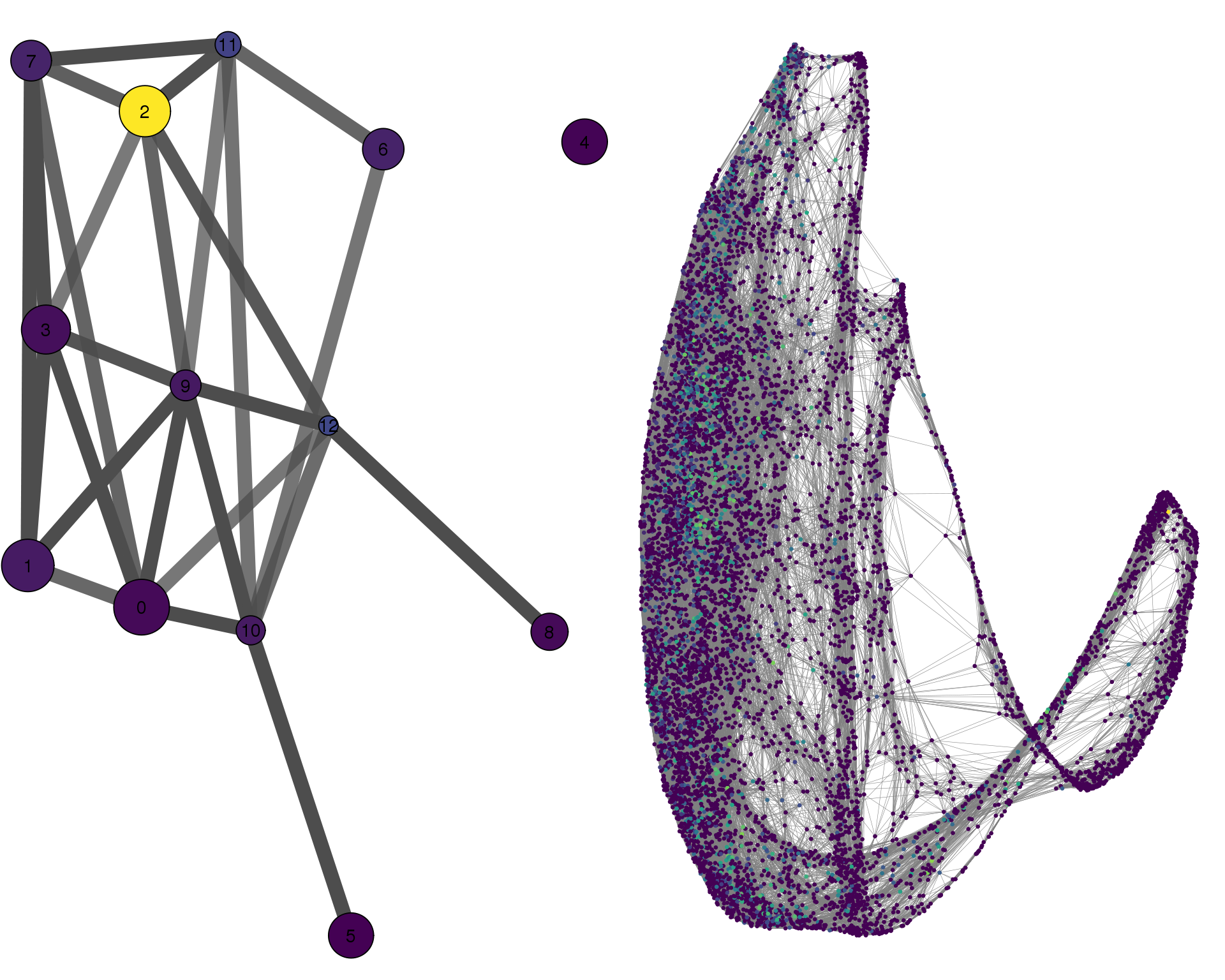
Expand here to see past versions of compare-CENPF-1.png:
| Version | Author | Date |
|---|---|---|
| fefdd07 | Luke Zappia | 2019-02-28 |
HMGB2
plotPAGACompare(clust_embedding, clust_edges,
clust_thresh = con_thresh, cell_embedding, cell_edges,
cell_thresh = 0.1, colour = 'HMGB2')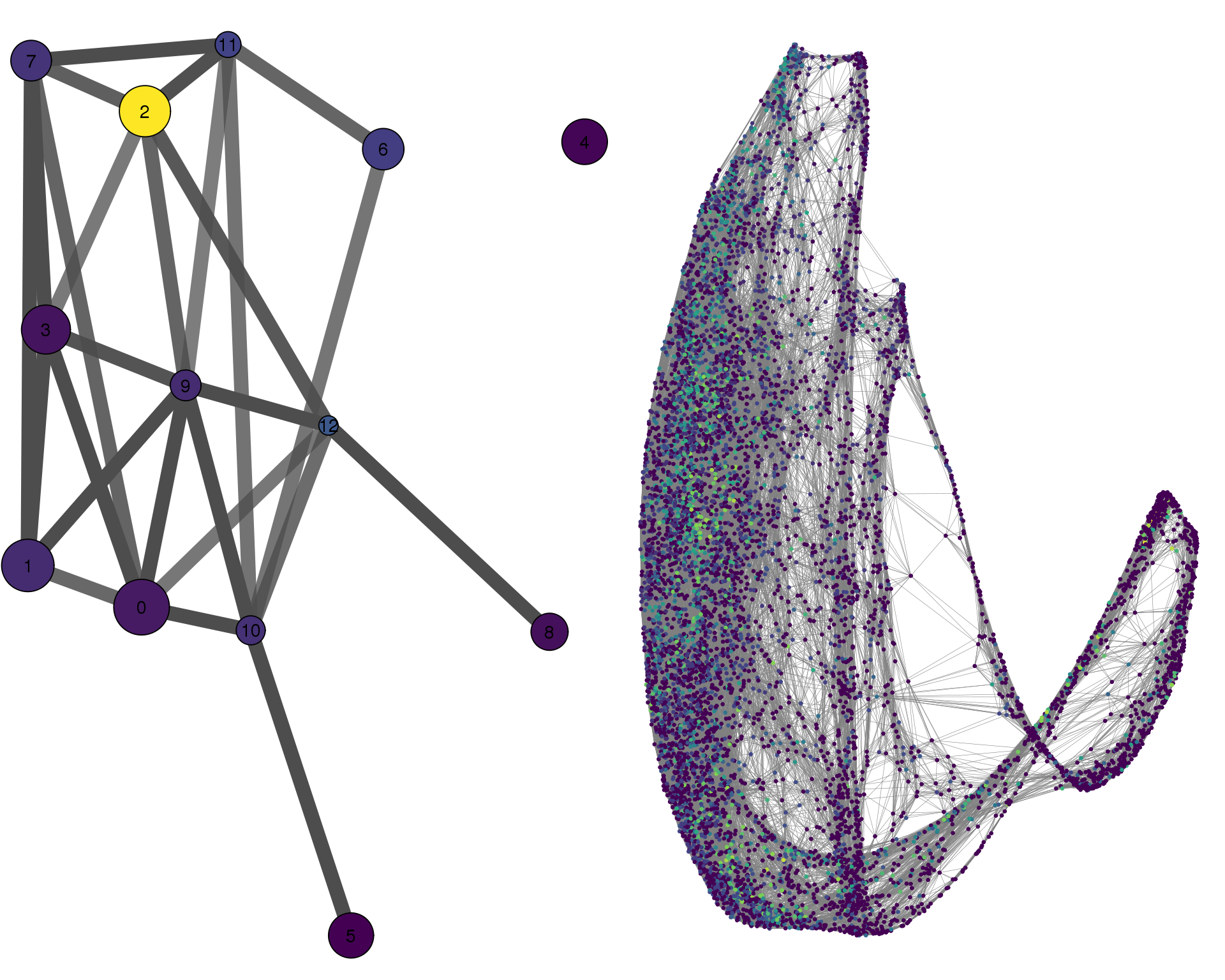
Expand here to see past versions of compare-HMGB2-1.png:
| Version | Author | Date |
|---|---|---|
| fefdd07 | Luke Zappia | 2019-02-28 |
CLDN5
plotPAGACompare(clust_embedding, clust_edges,
clust_thresh = con_thresh, cell_embedding, cell_edges,
cell_thresh = 0.1, colour = 'CLDN5')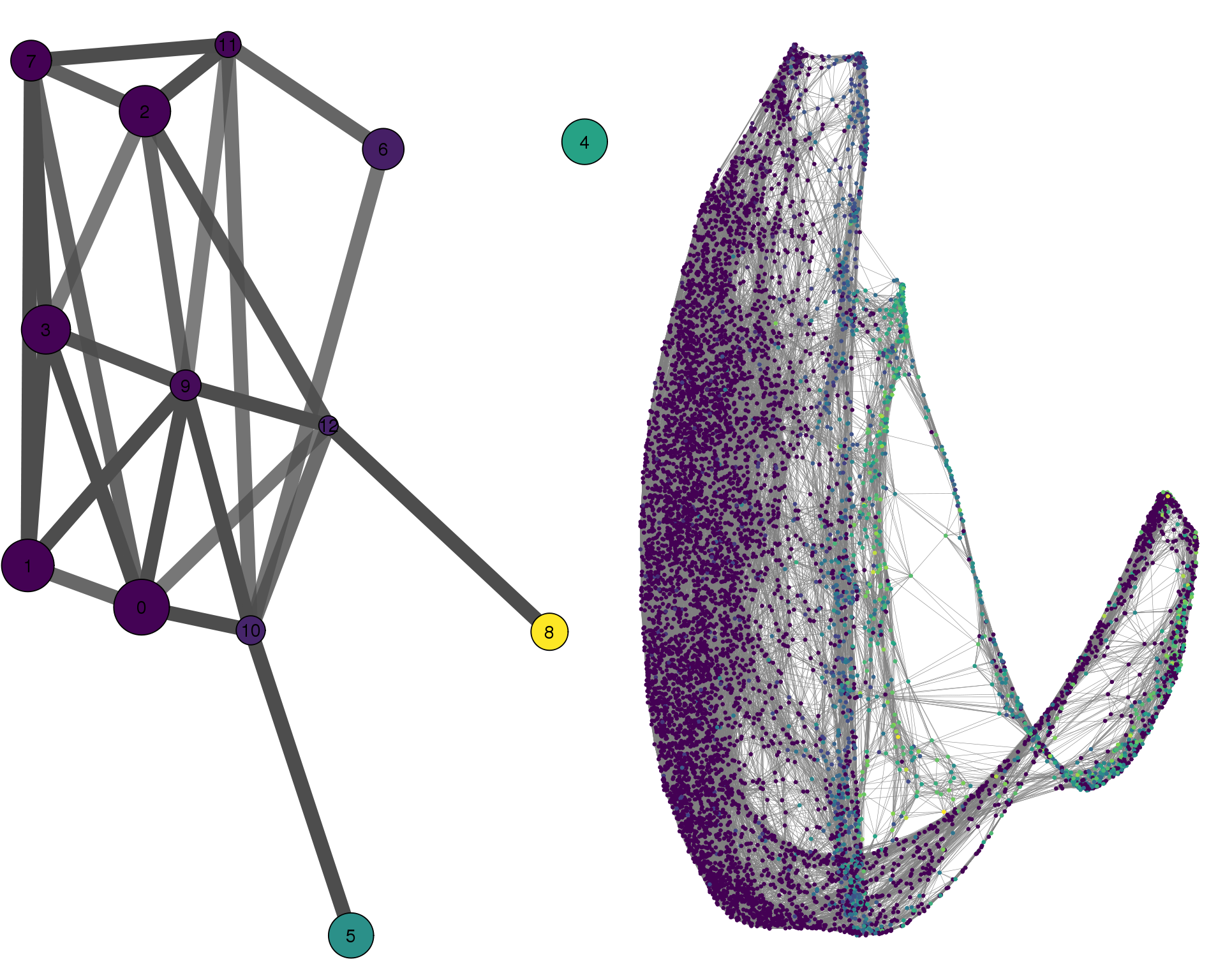
Expand here to see past versions of compare-CLDN5-1.png:
| Version | Author | Date |
|---|---|---|
| fefdd07 | Luke Zappia | 2019-02-28 |
PECAM1
plotPAGACompare(clust_embedding, clust_edges,
clust_thresh = con_thresh, cell_embedding, cell_edges,
cell_thresh = 0.1, colour = 'PECAM1')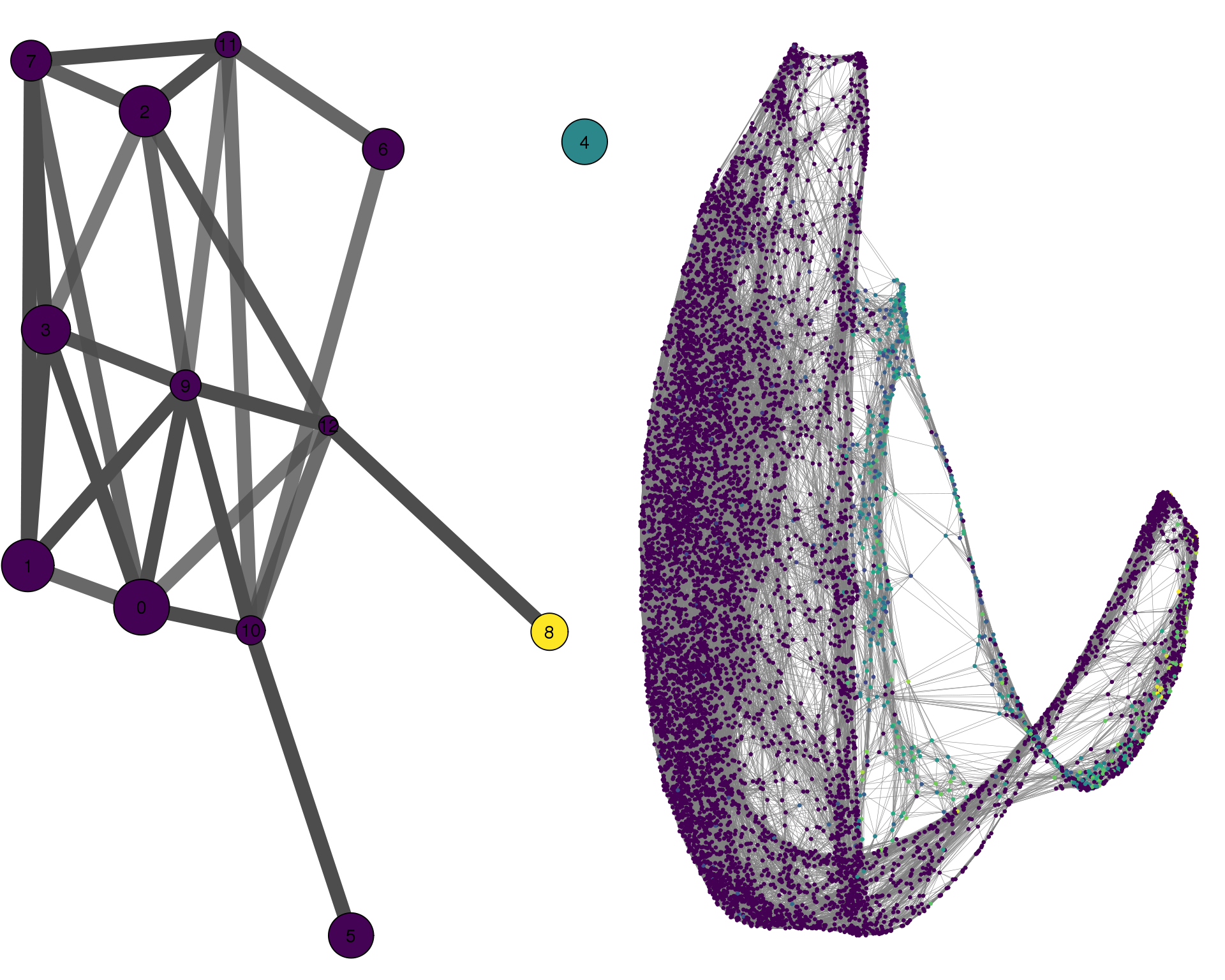
Expand here to see past versions of compare-PECAM1-1.png:
| Version | Author | Date |
|---|---|---|
| fefdd07 | Luke Zappia | 2019-02-28 |
KDR
plotPAGACompare(clust_embedding, clust_edges,
clust_thresh = con_thresh, cell_embedding, cell_edges,
cell_thresh = 0.1, colour = 'KDR')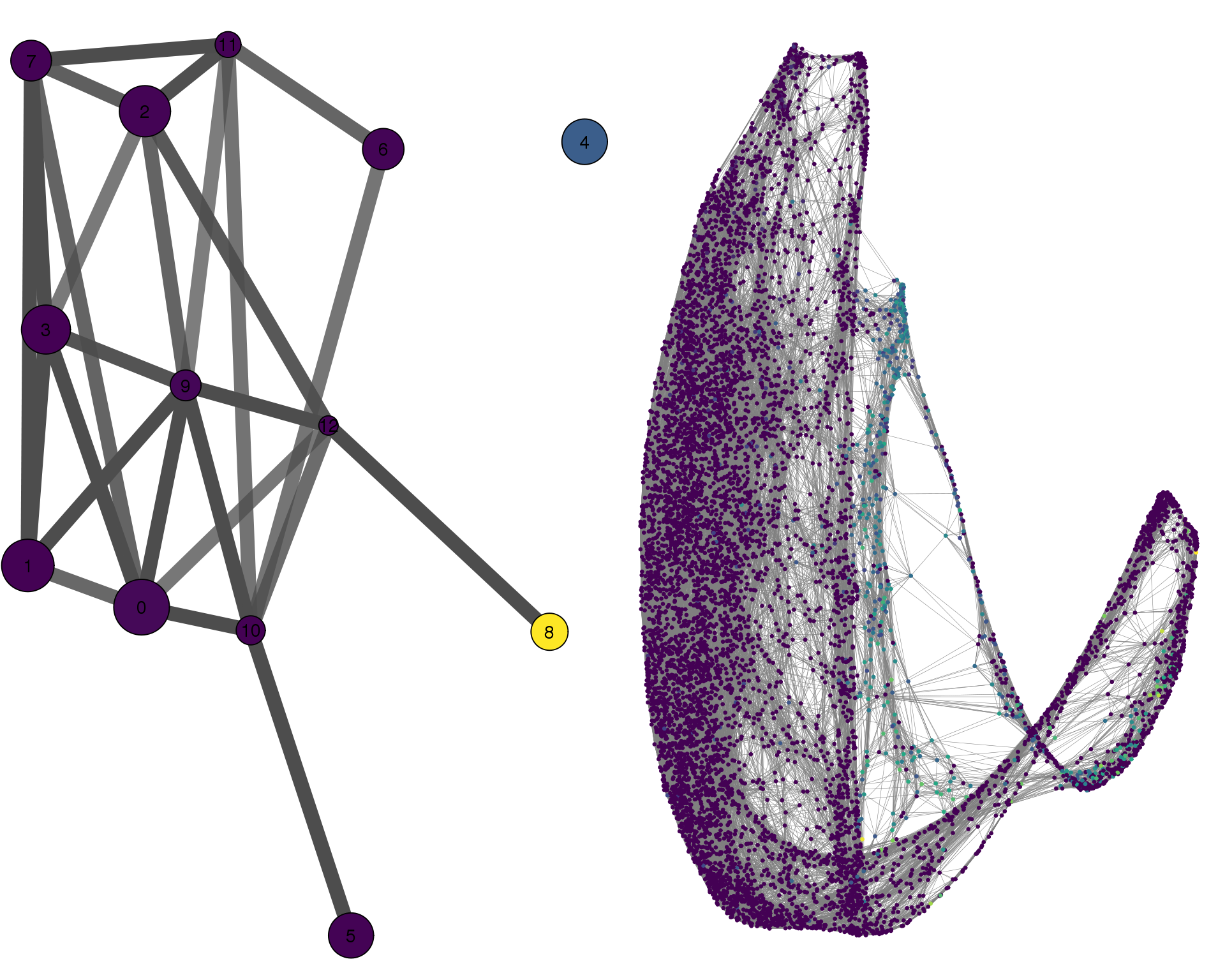
Expand here to see past versions of compare-KDR-1.png:
| Version | Author | Date |
|---|---|---|
| fefdd07 | Luke Zappia | 2019-02-28 |
CALM1
plotPAGACompare(clust_embedding, clust_edges,
clust_thresh = con_thresh, cell_embedding, cell_edges,
cell_thresh = 0.1, colour = 'CALM1')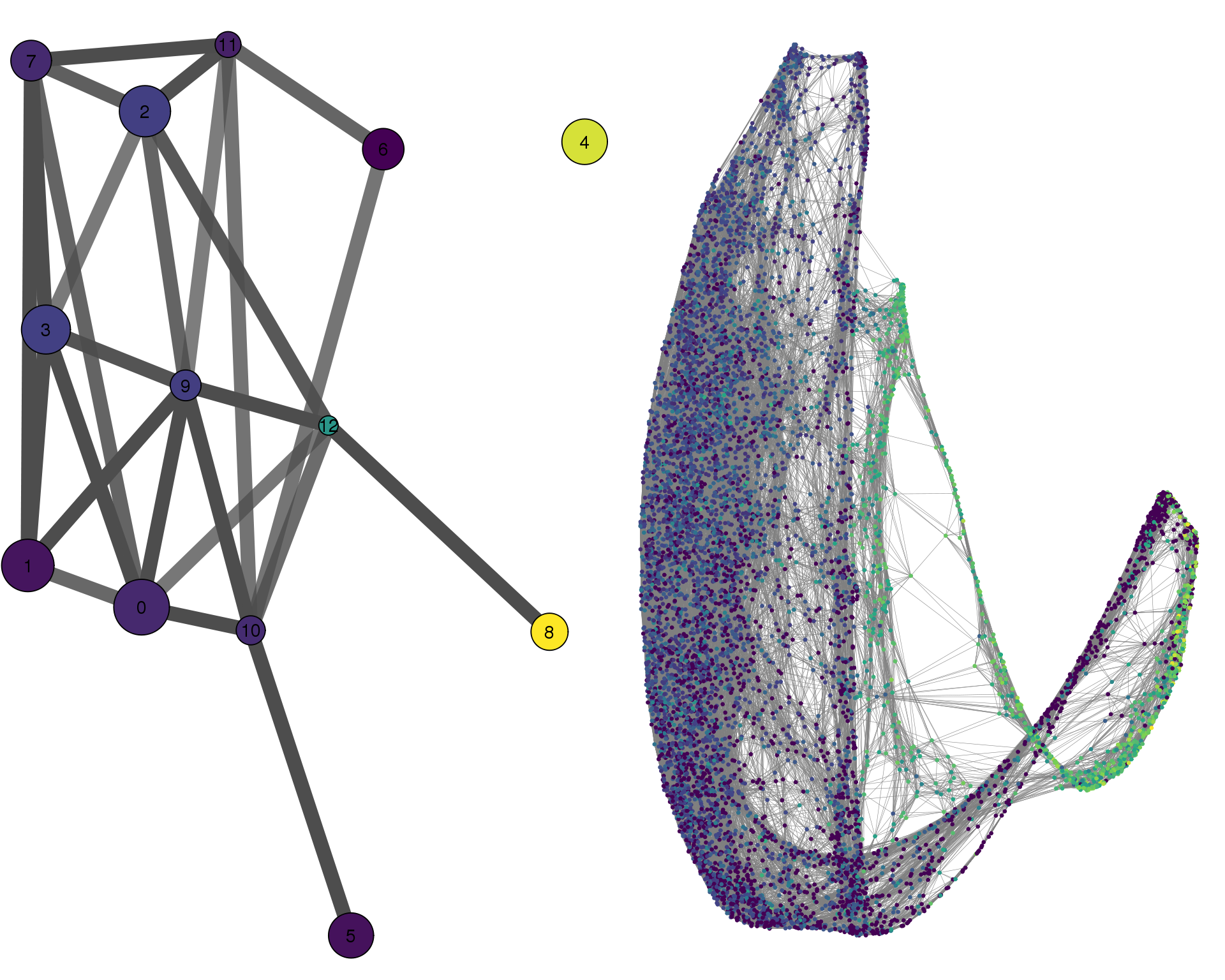
Expand here to see past versions of compare-CALM1-1.png:
| Version | Author | Date |
|---|---|---|
| fefdd07 | Luke Zappia | 2019-02-28 |
TTYH1
plotPAGACompare(clust_embedding, clust_edges,
clust_thresh = con_thresh, cell_embedding, cell_edges,
cell_thresh = 0.1, colour = 'TTYH1')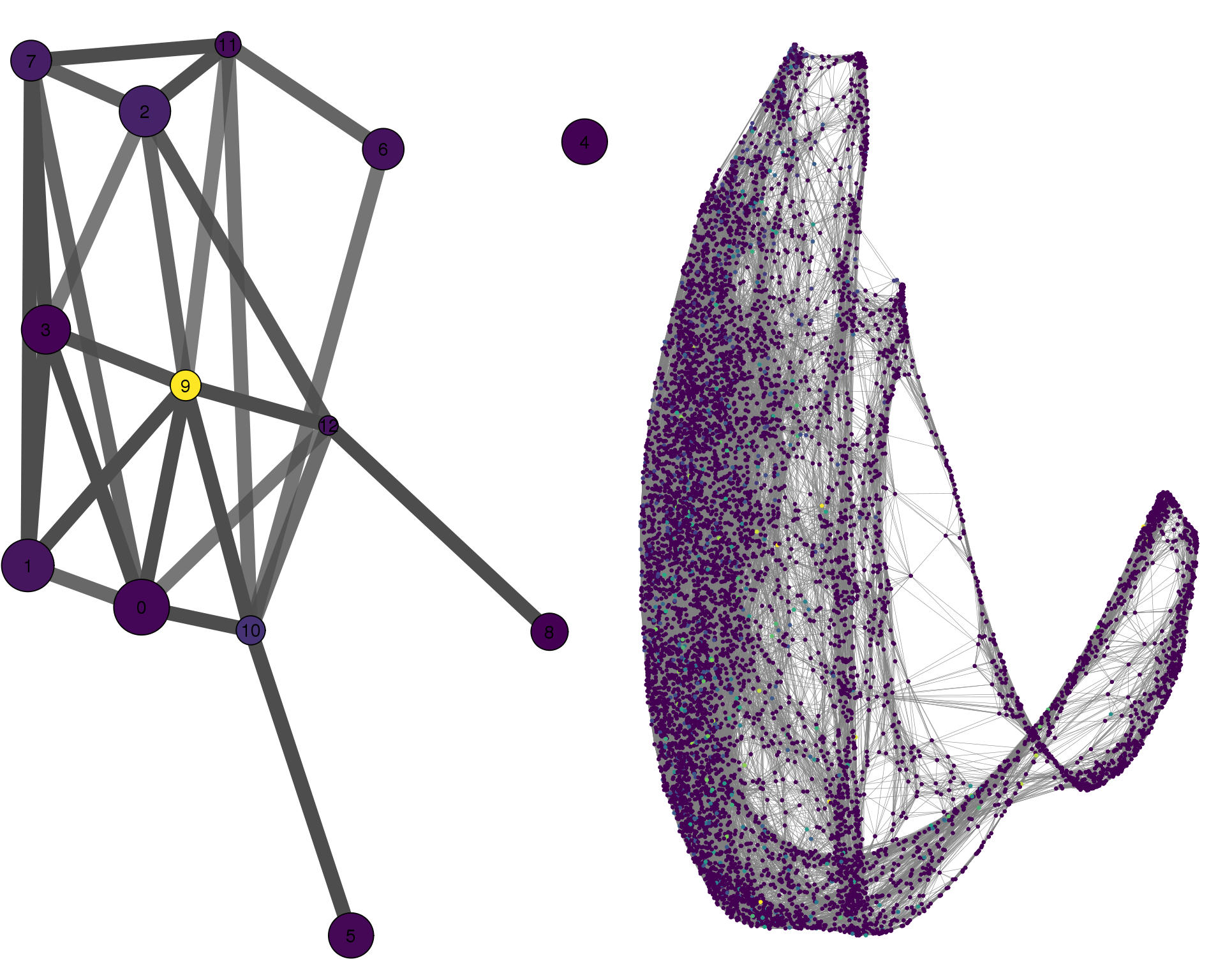
Expand here to see past versions of compare-TTYH1-1.png:
| Version | Author | Date |
|---|---|---|
| fefdd07 | Luke Zappia | 2019-02-28 |
SOX2
plotPAGACompare(clust_embedding, clust_edges,
clust_thresh = con_thresh, cell_embedding, cell_edges,
cell_thresh = 0.1, colour = 'SOX2')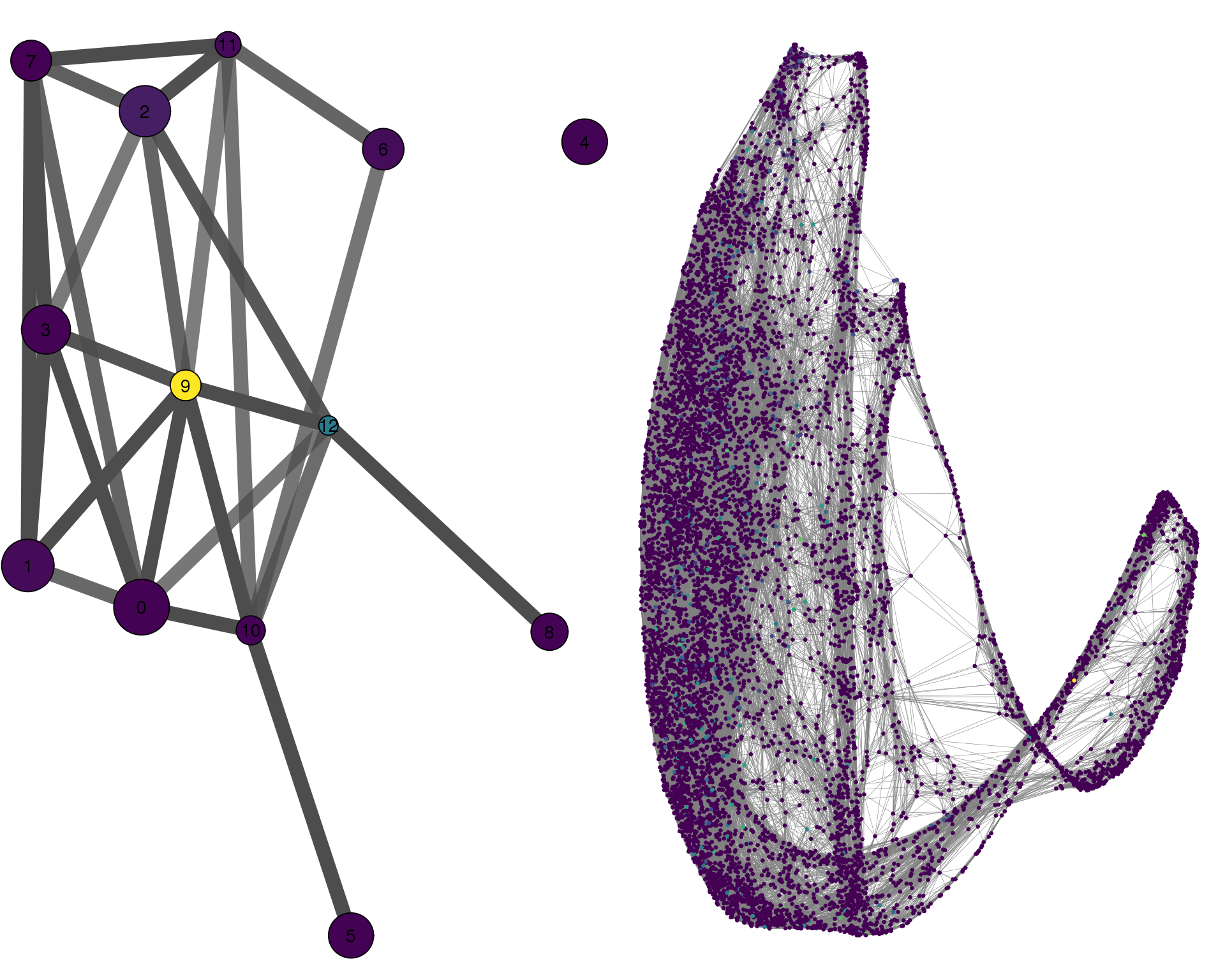
Expand here to see past versions of compare-SOX2-1.png:
| Version | Author | Date |
|---|---|---|
| fefdd07 | Luke Zappia | 2019-02-28 |
HES6
plotPAGACompare(clust_embedding, clust_edges,
clust_thresh = con_thresh, cell_embedding, cell_edges,
cell_thresh = 0.1, colour = 'HES6')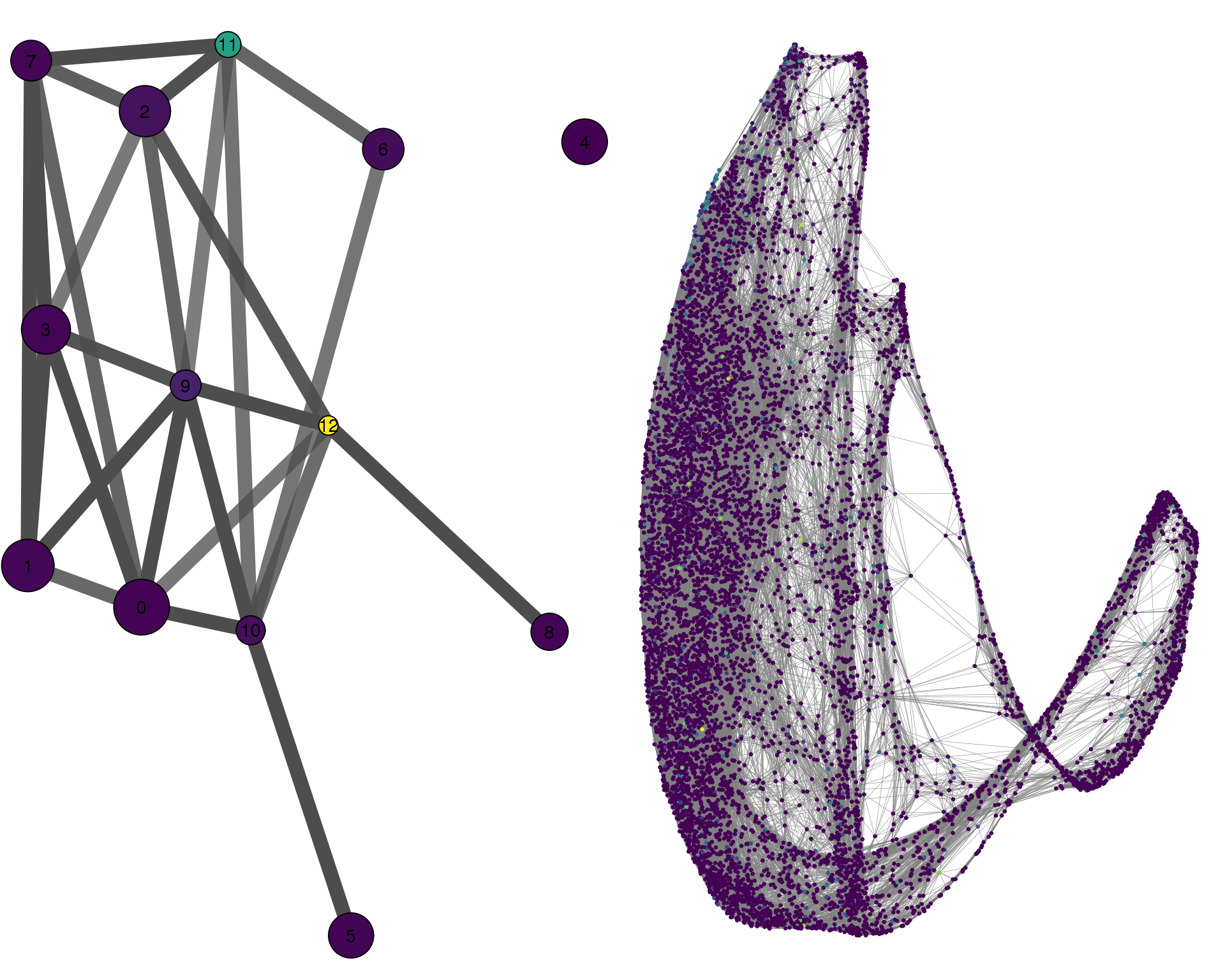
Expand here to see past versions of compare-HES6-1.png:
| Version | Author | Date |
|---|---|---|
| fefdd07 | Luke Zappia | 2019-02-28 |
STMN2
plotPAGACompare(clust_embedding, clust_edges,
clust_thresh = con_thresh, cell_embedding, cell_edges,
cell_thresh = 0.1, colour = 'STMN2')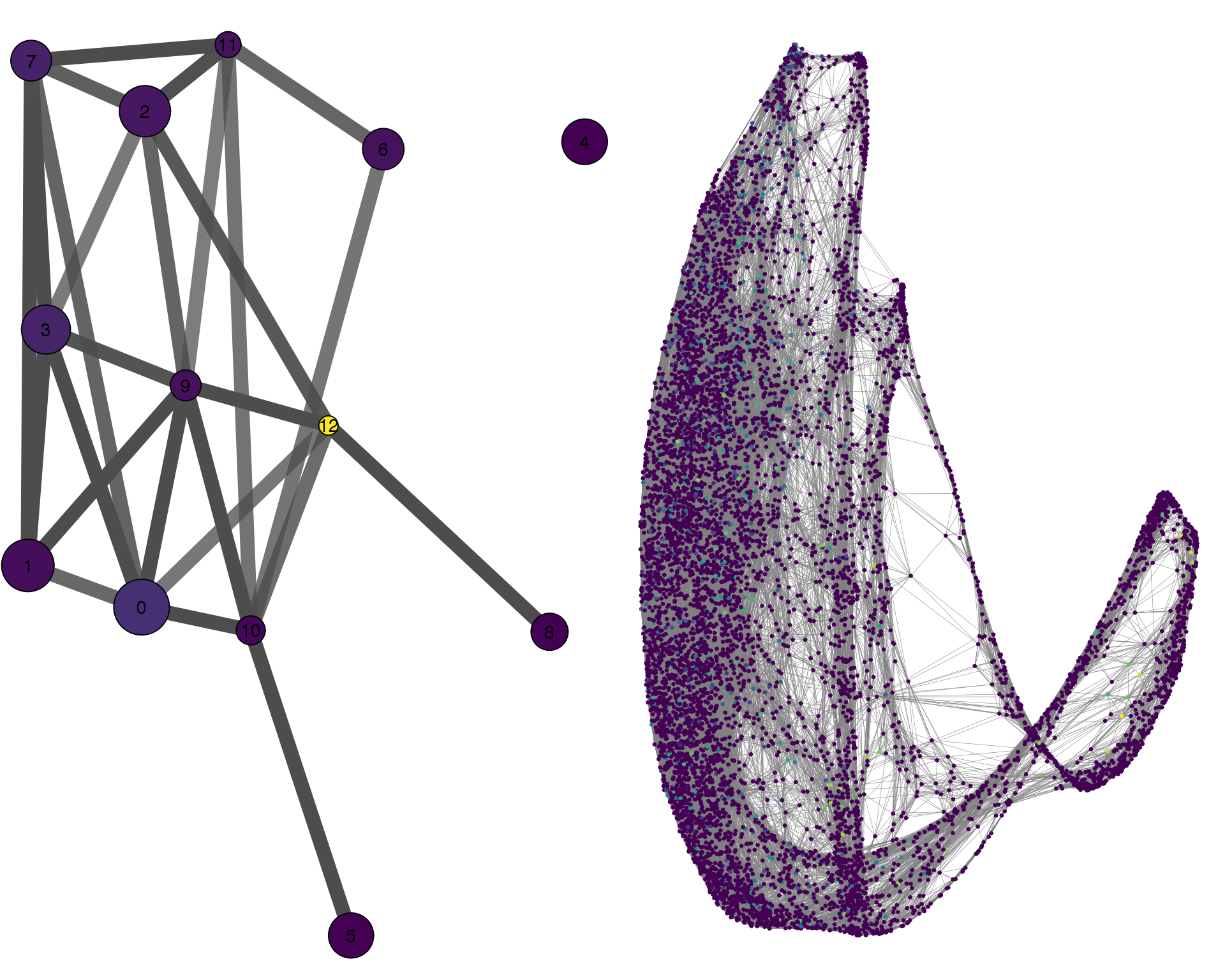
Expand here to see past versions of compare-STMN2-1.png:
| Version | Author | Date |
|---|---|---|
| fefdd07 | Luke Zappia | 2019-02-28 |
PAX2
plotPAGACompare(clust_embedding, clust_edges,
clust_thresh = con_thresh, cell_embedding, cell_edges,
cell_thresh = 0.1, colour = 'PAX2')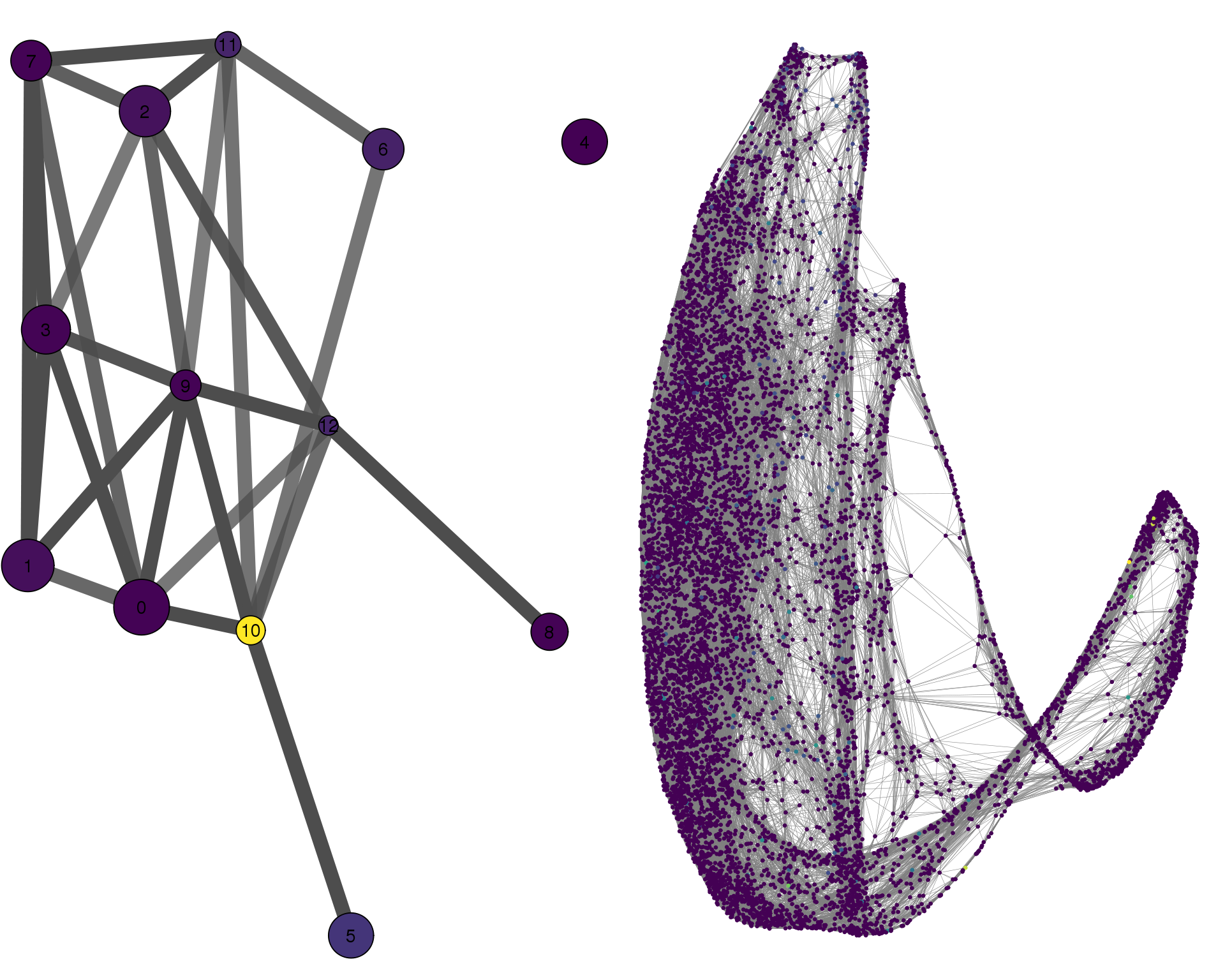
Expand here to see past versions of compare-PAX2-1.png:
| Version | Author | Date |
|---|---|---|
| fefdd07 | Luke Zappia | 2019-02-28 |
PAX8
plotPAGACompare(clust_embedding, clust_edges,
clust_thresh = con_thresh, cell_embedding, cell_edges,
cell_thresh = 0.1, colour = 'PAX8')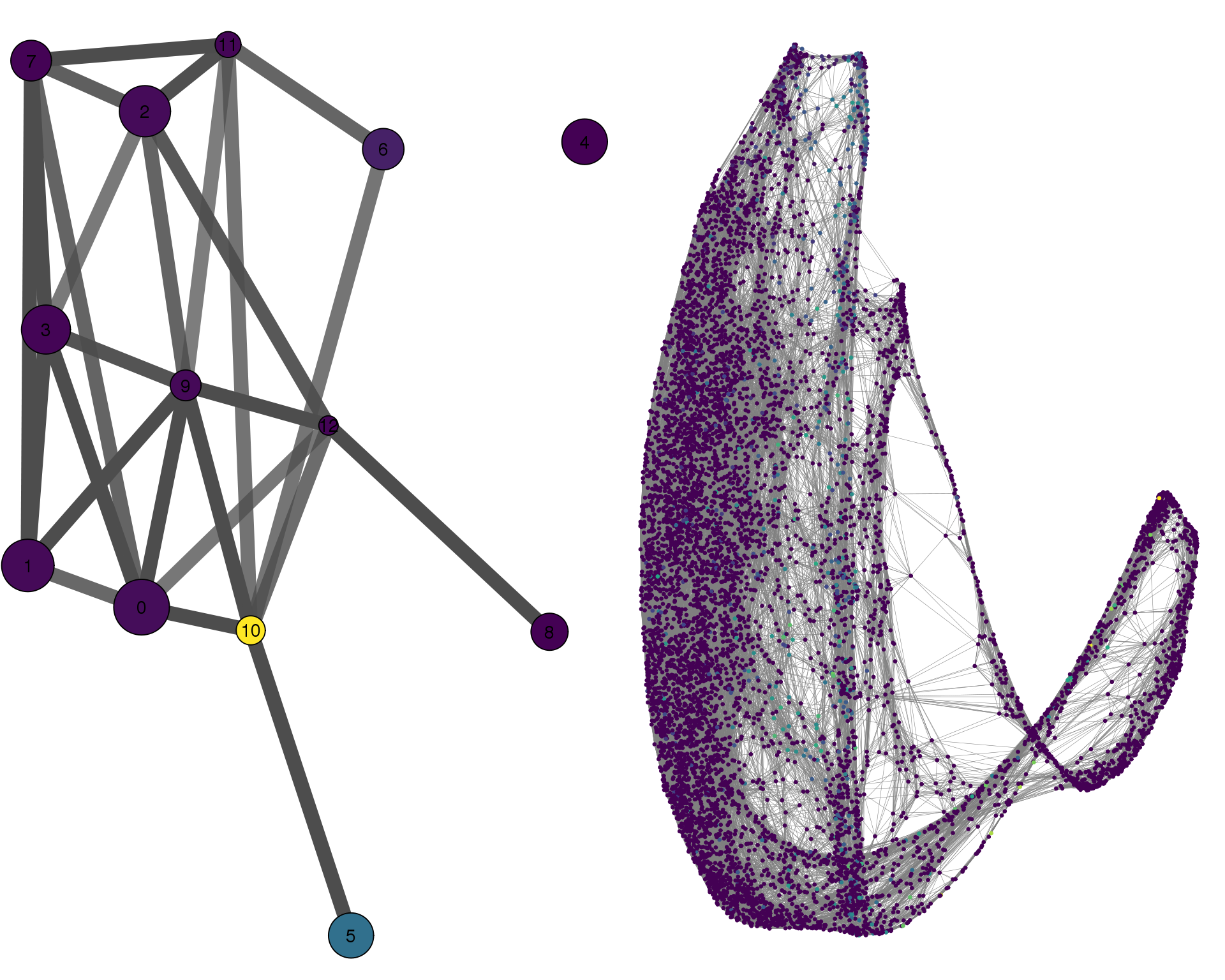
Expand here to see past versions of compare-PAX8-1.png:
| Version | Author | Date |
|---|---|---|
| fefdd07 | Luke Zappia | 2019-02-28 |
KRT19
plotPAGACompare(clust_embedding, clust_edges,
clust_thresh = con_thresh, cell_embedding, cell_edges,
cell_thresh = 0.1, colour = 'KRT19')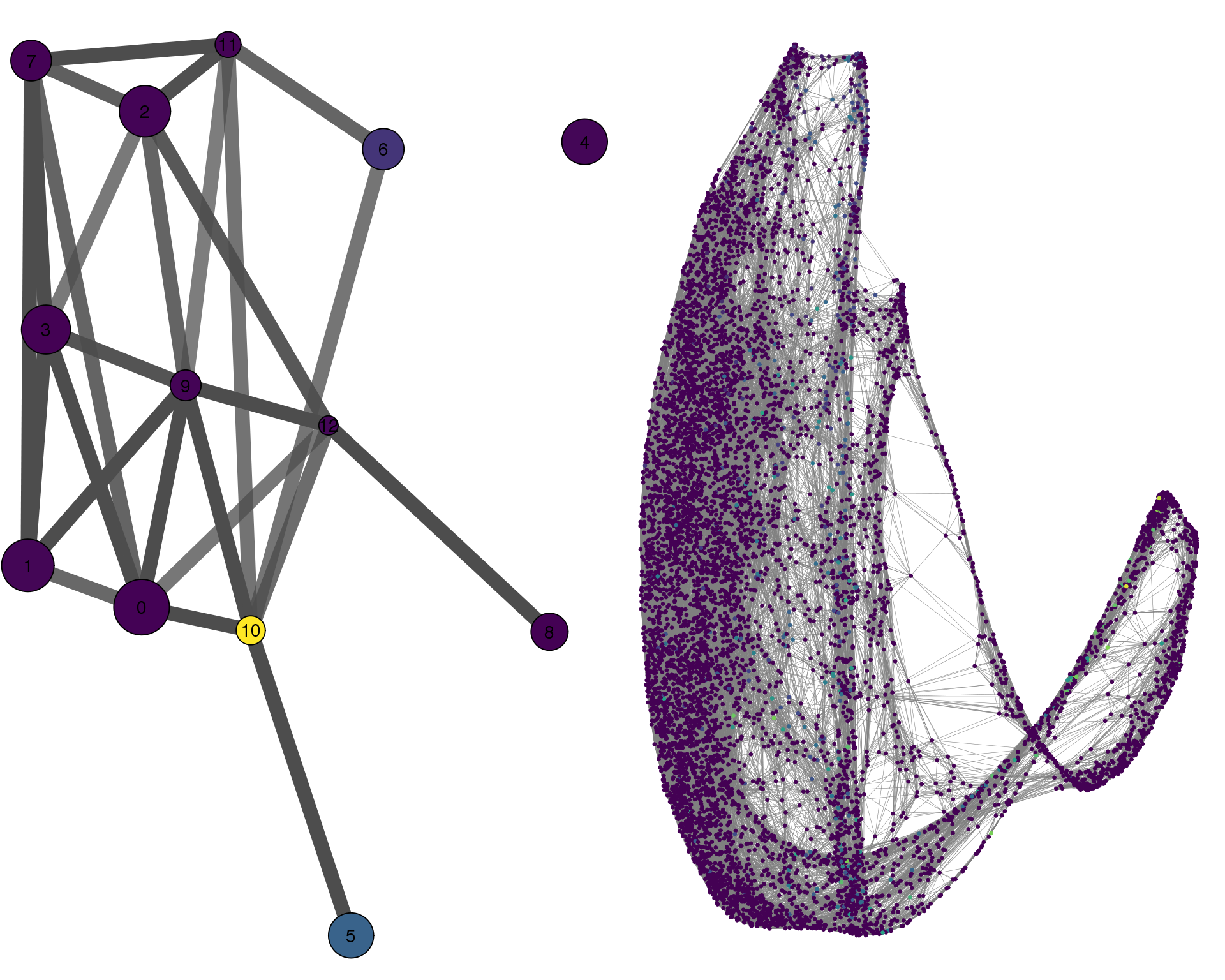
Expand here to see past versions of compare-KRT19-1.png:
| Version | Author | Date |
|---|---|---|
| fefdd07 | Luke Zappia | 2019-02-28 |
MYOG
plotPAGACompare(clust_embedding, clust_edges,
clust_thresh = con_thresh, cell_embedding, cell_edges,
cell_thresh = 0.1, colour = 'MYOG')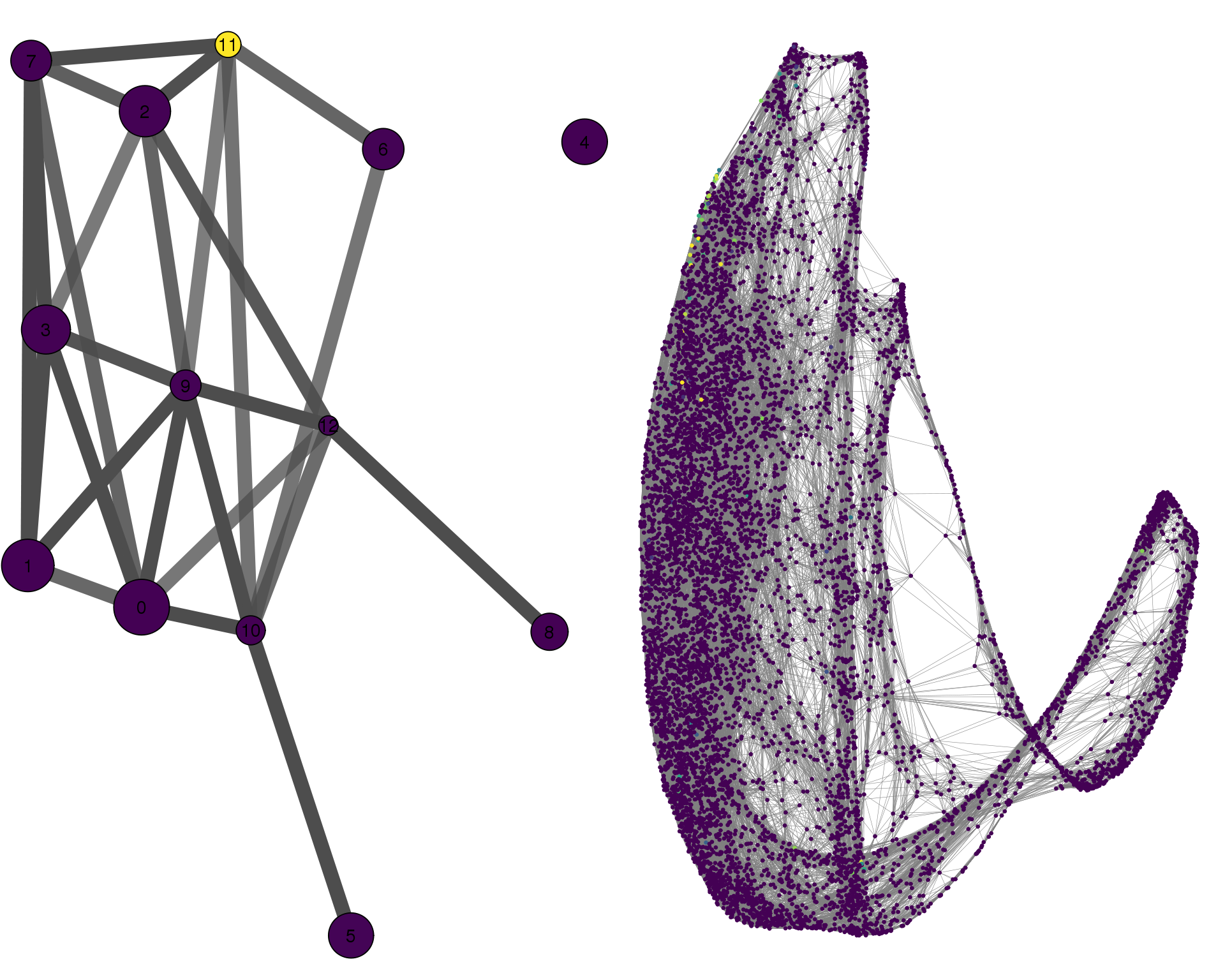
Expand here to see past versions of compare-MYOG-1.png:
| Version | Author | Date |
|---|---|---|
| fefdd07 | Luke Zappia | 2019-02-28 |
MYOD1
plotPAGACompare(clust_embedding, clust_edges,
clust_thresh = con_thresh, cell_embedding, cell_edges,
cell_thresh = 0.1, colour = 'MYOD1')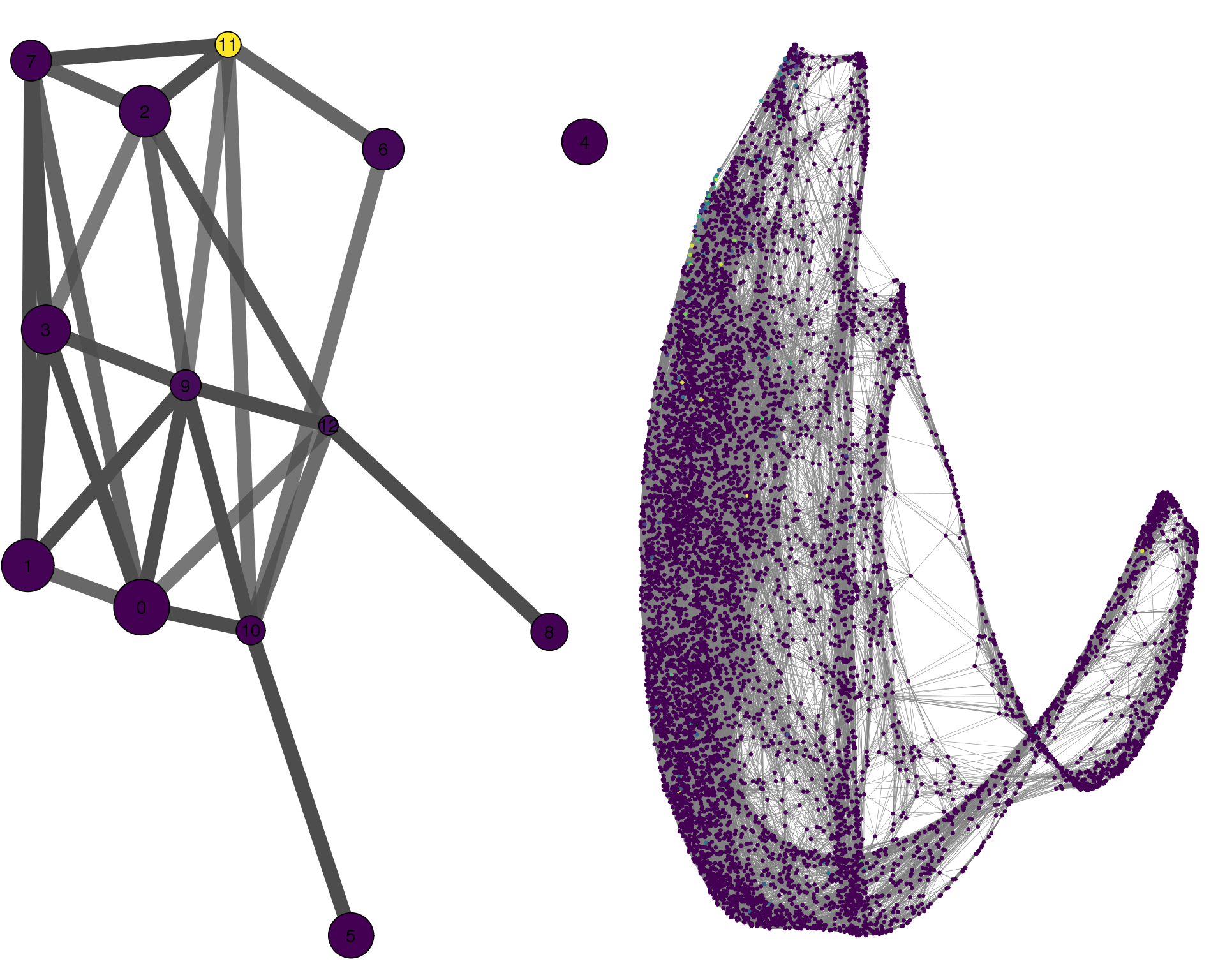
Expand here to see past versions of compare-MYOD1-1.png:
| Version | Author | Date |
|---|---|---|
| fefdd07 | Luke Zappia | 2019-02-28 |
Figures
clust_plot <- ggplot(clust_embedding, aes(x = X, y = Y)) +
geom_segment(data = clust_edges,
aes(x = FromX, y = FromY, xend = ToX, yend = ToY,
colour = Connectivity, alpha = Connectivity), size = 4) +
scale_colour_viridis_c(limits = c(0, 1)) +
scale_alpha_continuous(limits = c(0, 1), range = c(0, 1), guide = FALSE) +
geom_point(aes(fill = Cluster, size = Size), shape = 21) +
geom_text(aes(label = Cluster)) +
scale_size(range = c(6, 20), guide = FALSE) +
scale_fill_discrete(guide = FALSE) +
guides(colour = guide_colourbar(barwidth = 20)) +
ggtitle("PAGA cluster graph") +
theme_void() +
theme(plot.title = element_text(size = rel(1.2), hjust = 0.1,
vjust = 1, margin = margin(5)),
legend.position = "bottom")
cell_plot <- ggplot(cell_embedding, aes(x = X, y = Y)) +
geom_segment(data = cell_edges,
aes(x = FromX, y = FromY, xend = ToX, yend = ToY,
size = Connectivity, alpha = Connectivity)) +
geom_point(aes(colour = Cluster), size = 1) +
scale_alpha_continuous(limits = c(0, 1), range = c(0, 0.5), guide = FALSE) +
scale_colour_discrete(guide = guide_legend(
nrow = 1, override.aes = list(size = 4)
)) +
scale_size(range = c(0.1, 0.5), guide = FALSE) +
ggtitle("Cell shared nearest neighbour graph") +
theme_void() +
theme(plot.title = element_text(size = rel(1.2), hjust = 0.1,
vjust = 1, margin = margin(5)),
legend.position = "bottom")
fig <- plot_grid(clust_plot, cell_plot, nrow = 2, labels = "AUTO")
ggsave(here::here("output", DOCNAME, "paga-results.pdf"), fig,
width = 7, height = 8, scale = 1.5)
ggsave(here::here("output", DOCNAME, "paga-results.png"), fig,
width = 7, height = 8, scale = 1.5)
fig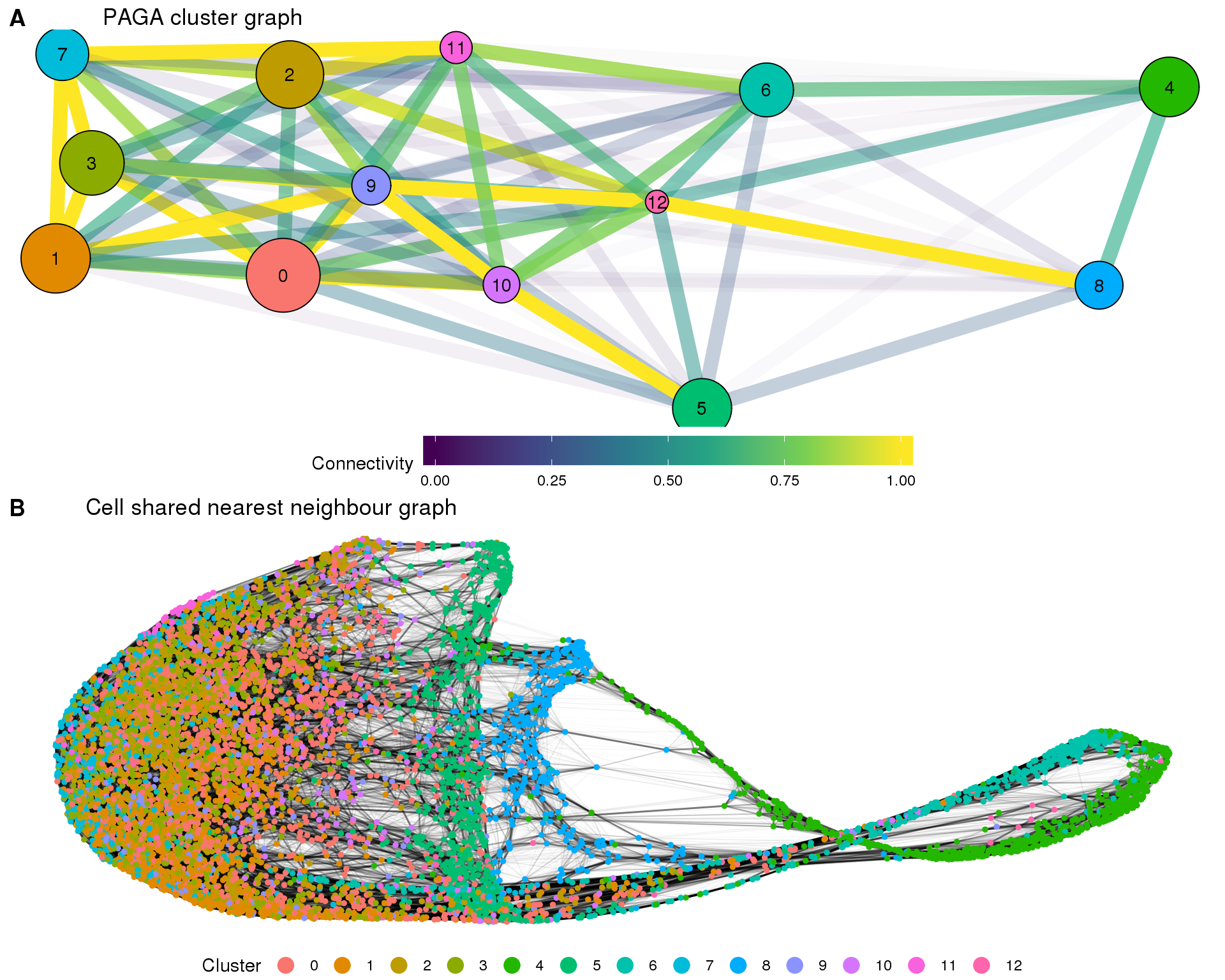
Summary
Parameters
This table describes parameters used and set in this document.
params <- list(
list(
Parameter = "con_thresh",
Value = con_thresh,
Description = "Connectivity threshold for PAGA graph"
)
)
params <- jsonlite::toJSON(params, pretty = TRUE)
knitr::kable(jsonlite::fromJSON(params))| Parameter | Value | Description |
|---|---|---|
| con_thresh | 0.7 | Connectivity threshold for PAGA graph |
Output files
This table describes the output files produced by this document. Right click and Save Link As… to download the results.
dir.create(here::here("output", DOCNAME), showWarnings = FALSE)
readr::write_lines(params, here::here("output", DOCNAME, "parameters.json"))
knitr::kable(data.frame(
File = c(
getDownloadLink("parameters.json", DOCNAME),
getDownloadLink("cluster_embedding.csv", DOCNAME),
getDownloadLink("cluster_edges.csv", DOCNAME),
getDownloadLink("cluster_tree_edges.csv", DOCNAME),
getDownloadLink("cell_embedding.csv", DOCNAME),
getDownloadLink("cell_edges.csv", DOCNAME),
getDownloadLink("paga-results.png", DOCNAME),
getDownloadLink("paga-results.pdf", DOCNAME)
),
Description = c(
"Parameters set and used in this analysis",
"Embedding for clusters from PAGA",
"Edges for PAGA cluster graph",
"Tree edges for PAGA cluster graph",
"Embedding for cells from PAGA",
"Edges for cell graph",
"PAGA results figure (PNG)",
"PAGA results figure (PDF)"
)
))| File | Description |
|---|---|
| parameters.json | Parameters set and used in this analysis |
| cluster_embedding.csv | Embedding for clusters from PAGA |
| cluster_edges.csv | Edges for PAGA cluster graph |
| cluster_tree_edges.csv | Tree edges for PAGA cluster graph |
| cell_embedding.csv | Embedding for cells from PAGA |
| cell_edges.csv | Edges for cell graph |
| paga-results.png | PAGA results figure (PNG) |
| paga-results.pdf | PAGA results figure (PDF) |
Session information
devtools::session_info()─ Session info ──────────────────────────────────────────────────────────
setting value
version R version 3.5.0 (2018-04-23)
os CentOS release 6.7 (Final)
system x86_64, linux-gnu
ui X11
language (EN)
collate en_US.UTF-8
ctype en_US.UTF-8
tz Australia/Melbourne
date 2019-04-03
─ Packages ──────────────────────────────────────────────────────────────
! package * version date lib source
assertthat 0.2.0 2017-04-11 [1] CRAN (R 3.5.0)
backports 1.1.3 2018-12-14 [1] CRAN (R 3.5.0)
bindr 0.1.1 2018-03-13 [1] CRAN (R 3.5.0)
bindrcpp 0.2.2 2018-03-29 [1] CRAN (R 3.5.0)
Biobase * 2.42.0 2018-10-30 [1] Bioconductor
BiocGenerics * 0.28.0 2018-10-30 [1] Bioconductor
BiocParallel * 1.16.5 2019-01-04 [1] Bioconductor
bitops 1.0-6 2013-08-17 [1] CRAN (R 3.5.0)
broom 0.5.1 2018-12-05 [1] CRAN (R 3.5.0)
callr 3.1.1 2018-12-21 [1] CRAN (R 3.5.0)
cellranger 1.1.0 2016-07-27 [1] CRAN (R 3.5.0)
cli 1.0.1 2018-09-25 [1] CRAN (R 3.5.0)
colorspace 1.4-0 2019-01-13 [1] CRAN (R 3.5.0)
cowplot * 0.9.4 2019-01-08 [1] CRAN (R 3.5.0)
crayon 1.3.4 2017-09-16 [1] CRAN (R 3.5.0)
DelayedArray * 0.8.0 2018-10-30 [1] Bioconductor
desc 1.2.0 2018-05-01 [1] CRAN (R 3.5.0)
devtools 2.0.1 2018-10-26 [1] CRAN (R 3.5.0)
digest 0.6.18 2018-10-10 [1] CRAN (R 3.5.0)
dplyr * 0.7.8 2018-11-10 [1] CRAN (R 3.5.0)
evaluate 0.12 2018-10-09 [1] CRAN (R 3.5.0)
forcats * 0.3.0 2018-02-19 [1] CRAN (R 3.5.0)
fs 1.2.6 2018-08-23 [1] CRAN (R 3.5.0)
generics 0.0.2 2018-11-29 [1] CRAN (R 3.5.0)
GenomeInfoDb * 1.18.1 2018-11-12 [1] Bioconductor
GenomeInfoDbData 1.2.0 2019-01-15 [1] Bioconductor
GenomicRanges * 1.34.0 2018-10-30 [1] Bioconductor
ggplot2 * 3.1.0 2018-10-25 [1] CRAN (R 3.5.0)
git2r 0.24.0 2019-01-07 [1] CRAN (R 3.5.0)
glue 1.3.0 2018-07-17 [1] CRAN (R 3.5.0)
gtable 0.2.0 2016-02-26 [1] CRAN (R 3.5.0)
haven 2.0.0 2018-11-22 [1] CRAN (R 3.5.0)
here 0.1 2017-05-28 [1] CRAN (R 3.5.0)
hms 0.4.2 2018-03-10 [1] CRAN (R 3.5.0)
htmltools 0.3.6 2017-04-28 [1] CRAN (R 3.5.0)
httr 1.4.0 2018-12-11 [1] CRAN (R 3.5.0)
IRanges * 2.16.0 2018-10-30 [1] Bioconductor
jsonlite 1.6 2018-12-07 [1] CRAN (R 3.5.0)
knitr * 1.21 2018-12-10 [1] CRAN (R 3.5.0)
P lattice 0.20-35 2017-03-25 [5] CRAN (R 3.5.0)
lazyeval 0.2.1 2017-10-29 [1] CRAN (R 3.5.0)
lubridate 1.7.4 2018-04-11 [1] CRAN (R 3.5.0)
magrittr 1.5 2014-11-22 [1] CRAN (R 3.5.0)
P Matrix 1.2-14 2018-04-09 [5] CRAN (R 3.5.0)
matrixStats * 0.54.0 2018-07-23 [1] CRAN (R 3.5.0)
memoise 1.1.0 2017-04-21 [1] CRAN (R 3.5.0)
modelr 0.1.3 2019-02-05 [1] CRAN (R 3.5.0)
munsell 0.5.0 2018-06-12 [1] CRAN (R 3.5.0)
P nlme 3.1-137 2018-04-07 [5] CRAN (R 3.5.0)
pillar 1.3.1 2018-12-15 [1] CRAN (R 3.5.0)
pkgbuild 1.0.2 2018-10-16 [1] CRAN (R 3.5.0)
pkgconfig 2.0.2 2018-08-16 [1] CRAN (R 3.5.0)
pkgload 1.0.2 2018-10-29 [1] CRAN (R 3.5.0)
plyr 1.8.4 2016-06-08 [1] CRAN (R 3.5.0)
prettyunits 1.0.2 2015-07-13 [1] CRAN (R 3.5.0)
processx * 3.2.1 2018-12-05 [1] CRAN (R 3.5.0)
ps 1.3.0 2018-12-21 [1] CRAN (R 3.5.0)
purrr * 0.3.0 2019-01-27 [1] CRAN (R 3.5.0)
R.methodsS3 1.7.1 2016-02-16 [1] CRAN (R 3.5.0)
R.oo 1.22.0 2018-04-22 [1] CRAN (R 3.5.0)
R.utils 2.7.0 2018-08-27 [1] CRAN (R 3.5.0)
R6 2.3.0 2018-10-04 [1] CRAN (R 3.5.0)
Rcpp 1.0.0 2018-11-07 [1] CRAN (R 3.5.0)
RCurl 1.95-4.11 2018-07-15 [1] CRAN (R 3.5.0)
readr * 1.3.1 2018-12-21 [1] CRAN (R 3.5.0)
readxl 1.2.0 2018-12-19 [1] CRAN (R 3.5.0)
remotes 2.0.2 2018-10-30 [1] CRAN (R 3.5.0)
rlang 0.3.1 2019-01-08 [1] CRAN (R 3.5.0)
rmarkdown 1.11 2018-12-08 [1] CRAN (R 3.5.0)
rprojroot 1.3-2 2018-01-03 [1] CRAN (R 3.5.0)
rstudioapi 0.9.0 2019-01-09 [1] CRAN (R 3.5.0)
rvest 0.3.2 2016-06-17 [1] CRAN (R 3.5.0)
S4Vectors * 0.20.1 2018-11-09 [1] Bioconductor
scales 1.0.0 2018-08-09 [1] CRAN (R 3.5.0)
sessioninfo 1.1.1 2018-11-05 [1] CRAN (R 3.5.0)
SingleCellExperiment * 1.4.1 2019-01-04 [1] Bioconductor
stringi 1.2.4 2018-07-20 [1] CRAN (R 3.5.0)
stringr * 1.3.1 2018-05-10 [1] CRAN (R 3.5.0)
SummarizedExperiment * 1.12.0 2018-10-30 [1] Bioconductor
testthat 2.0.1 2018-10-13 [4] CRAN (R 3.5.0)
tibble * 2.0.1 2019-01-12 [1] CRAN (R 3.5.0)
tidyr * 0.8.2 2018-10-28 [1] CRAN (R 3.5.0)
tidyselect 0.2.5 2018-10-11 [1] CRAN (R 3.5.0)
tidyverse * 1.2.1 2017-11-14 [1] CRAN (R 3.5.0)
usethis 1.4.0 2018-08-14 [1] CRAN (R 3.5.0)
whisker 0.3-2 2013-04-28 [1] CRAN (R 3.5.0)
withr 2.1.2 2018-03-15 [1] CRAN (R 3.5.0)
workflowr 1.1.1 2018-07-06 [1] CRAN (R 3.5.0)
xfun 0.4 2018-10-23 [1] CRAN (R 3.5.0)
xml2 1.2.0 2018-01-24 [1] CRAN (R 3.5.0)
XVector 0.22.0 2018-10-30 [1] Bioconductor
yaml 2.2.0 2018-07-25 [1] CRAN (R 3.5.0)
zlibbioc 1.28.0 2018-10-30 [1] Bioconductor
[1] /group/bioi1/luke/analysis/phd-thesis-analysis/packrat/lib/x86_64-pc-linux-gnu/3.5.0
[2] /group/bioi1/luke/analysis/phd-thesis-analysis/packrat/lib-ext/x86_64-pc-linux-gnu/3.5.0
[3] /group/bioi1/luke/analysis/phd-thesis-analysis/packrat/lib-R/x86_64-pc-linux-gnu/3.5.0
[4] /home/luke.zappia/R/x86_64-pc-linux-gnu-library/3.5
[5] /usr/local/installed/R/3.5.0/lib64/R/library
P ── Loaded and on-disk path mismatch.This reproducible R Markdown analysis was created with workflowr 1.1.1