gi2020
Genome Informatics Conference 2020
Last built: 2020-09-17 10:04:18
| Parameter | Value |
|---|---|
| hashtag | #gi2020 |
| start_day | 2020-09-14 |
| end_day | 2020-09-16 |
| timezone | Europe/London |
| theme | theme_light |
| accent | #e75152 |
| accent2 | #F3A8A8 |
| kcore | 2 |
| topics_k | 6 |
| bigram_filter | 3 |
| fixed | TRUE |
| seed | 1 |
Introduction
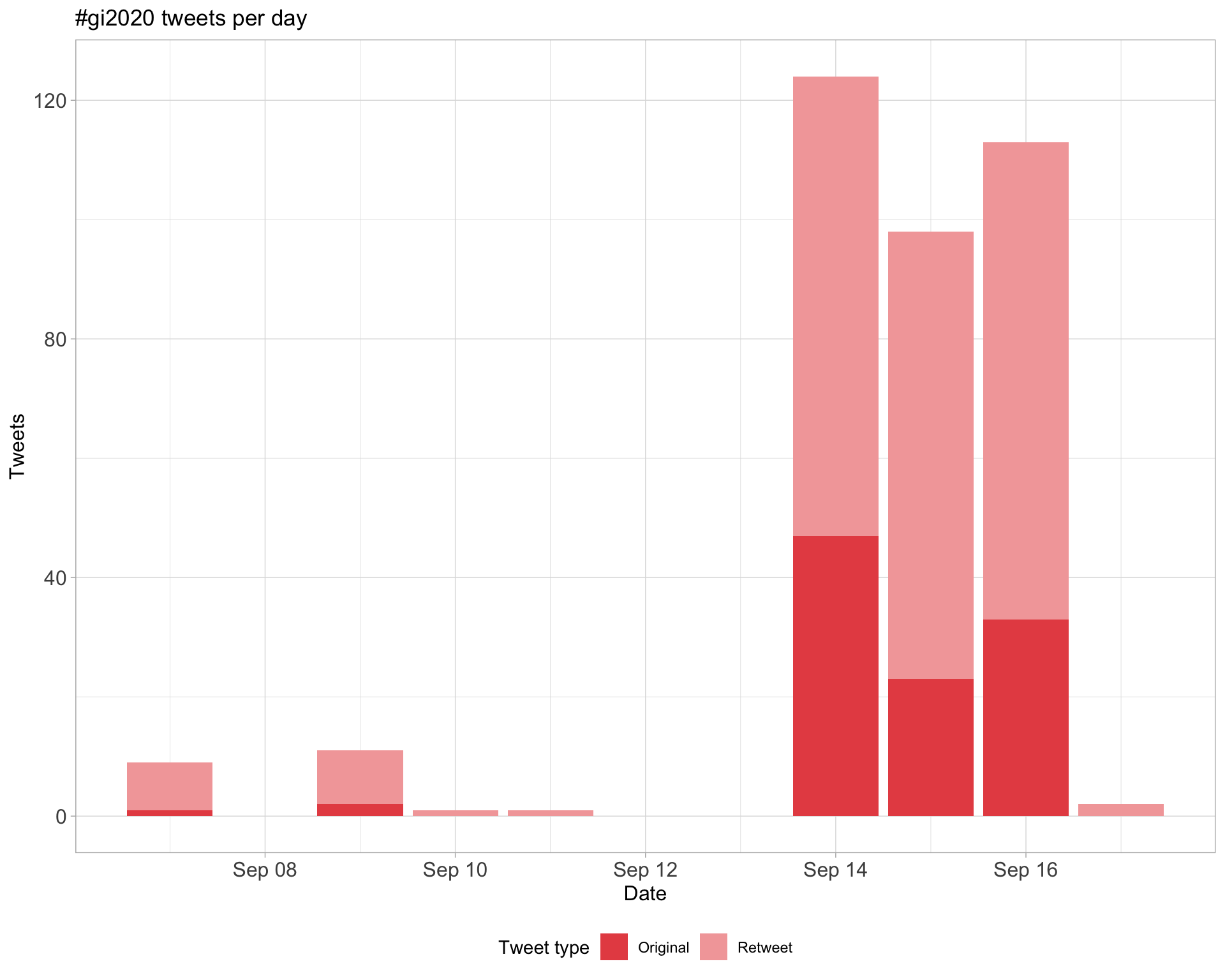
An analysis of tweets for the query #gi2020 related to Genome Informatics Conference 2020 from 2020-09-14 to 2020-09-16 (online due to the COVID-19 pandemic).
A total of 359 tweets from 203 users were collected using the {rtweet} R package.
1 Timeline
1.1 Tweets by day
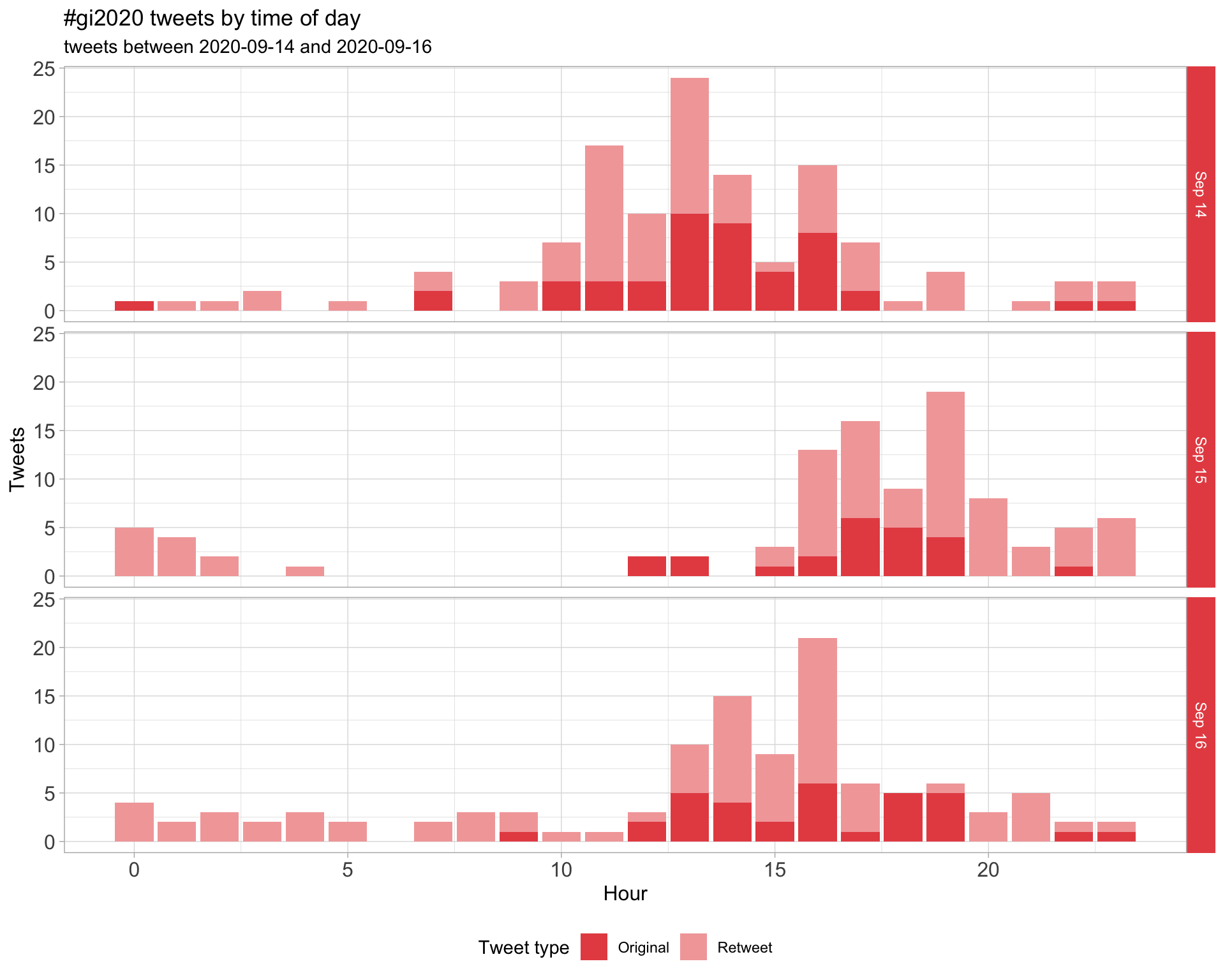
1.2 Tweets by day and time
Filtered for dates 2020-09-14 - 2020-09-16 in the Europe/London timezone.
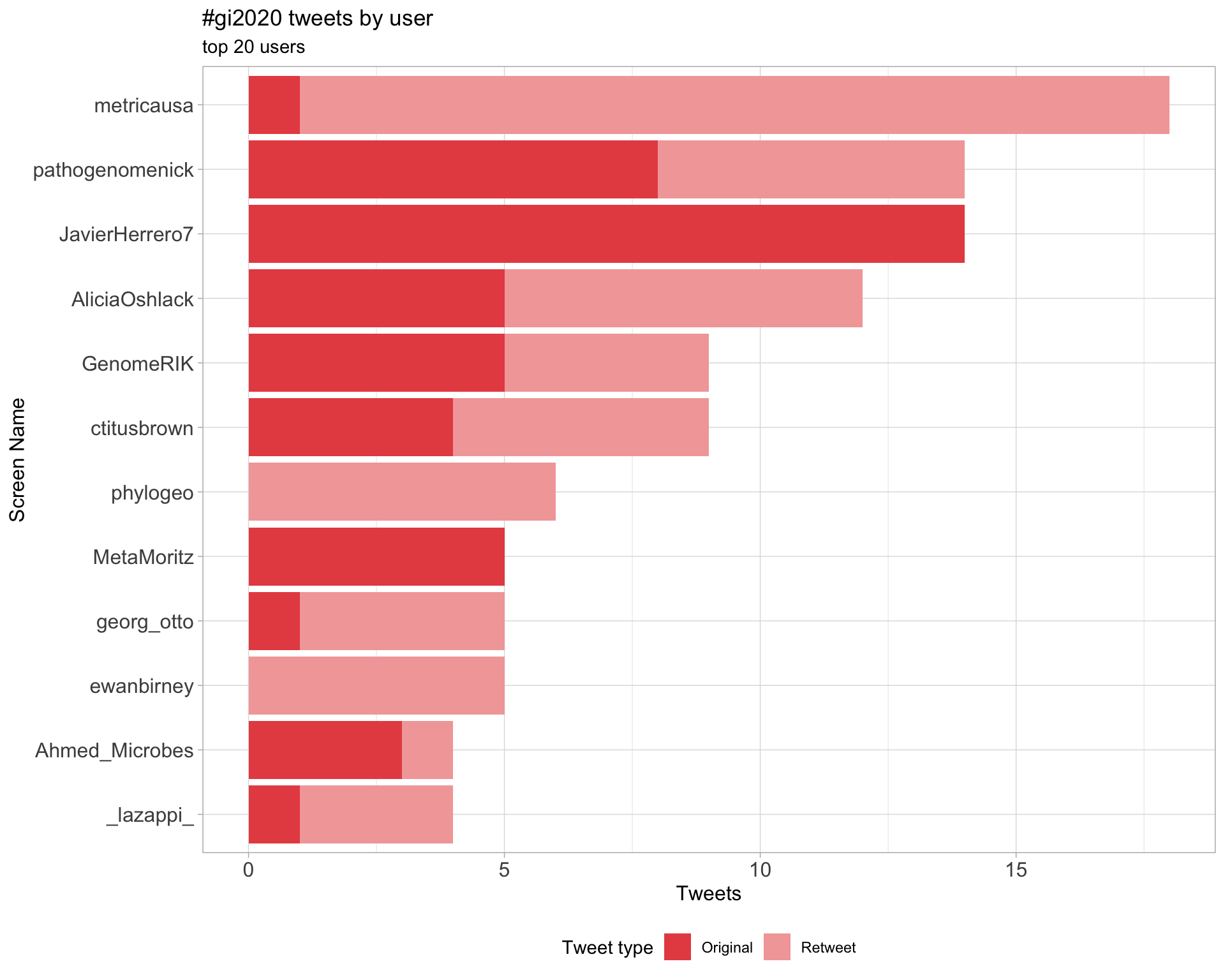
2 Users
2.1 Top tweeters
Overall
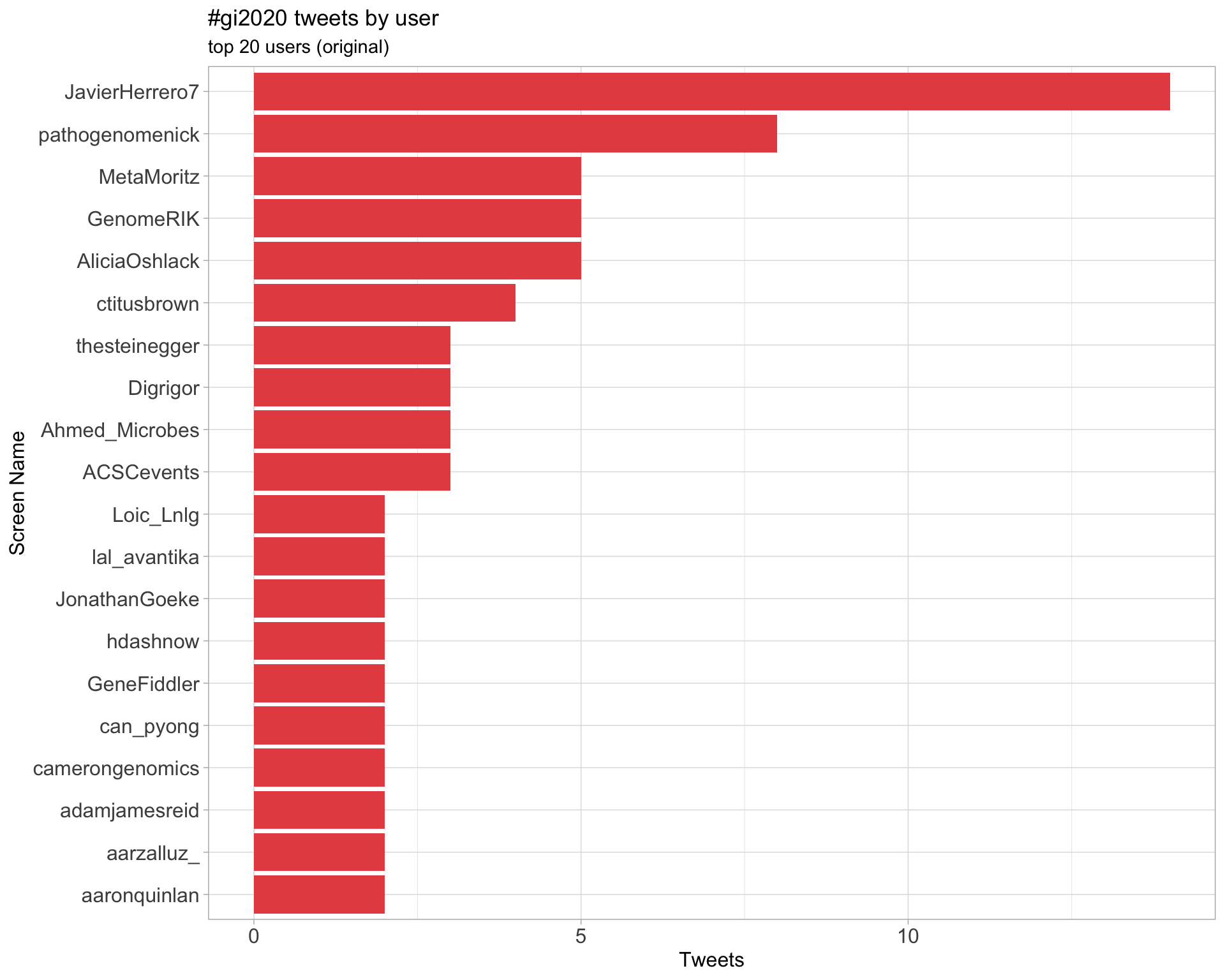
Original
Retweets
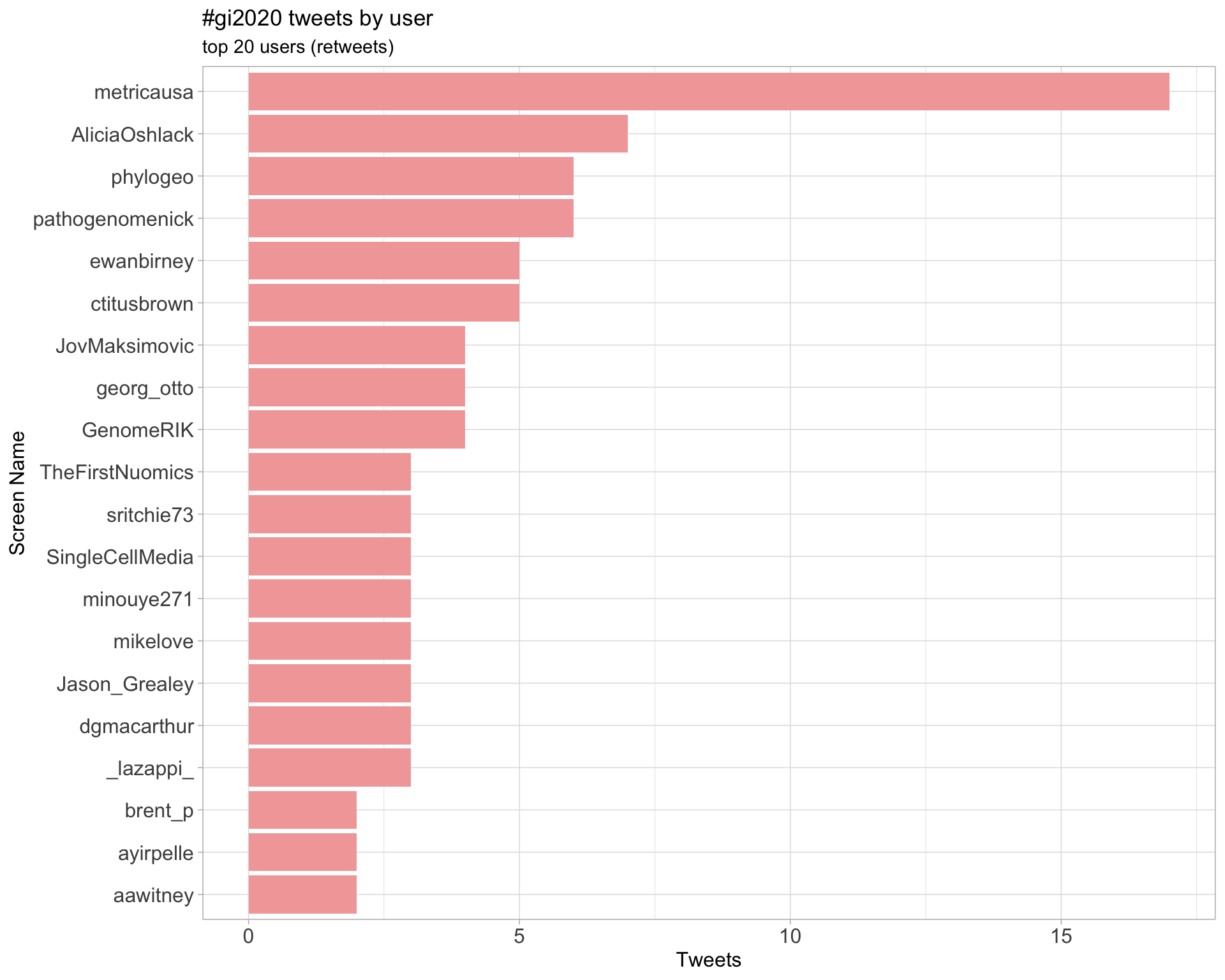
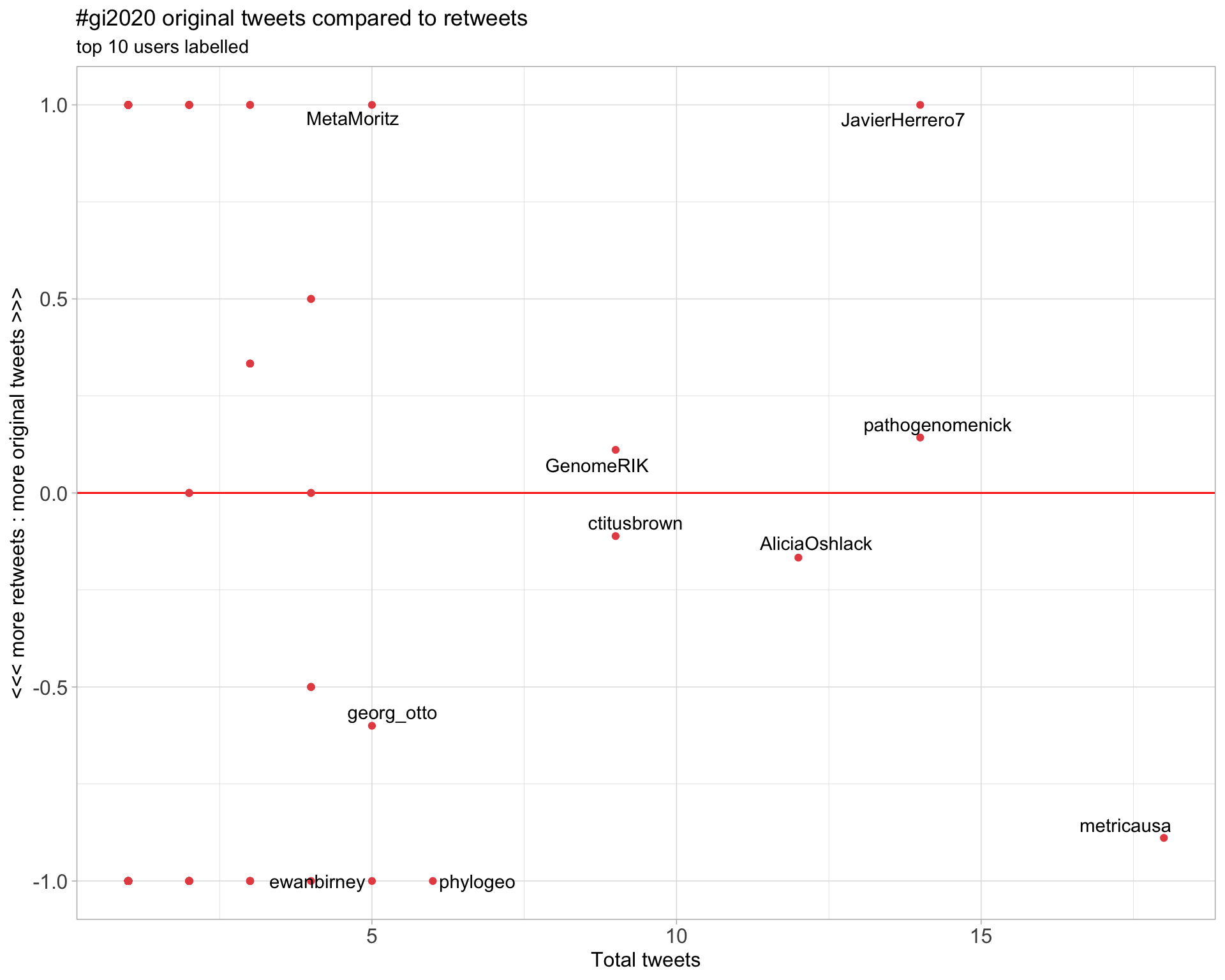
2.2 Retweet proportion
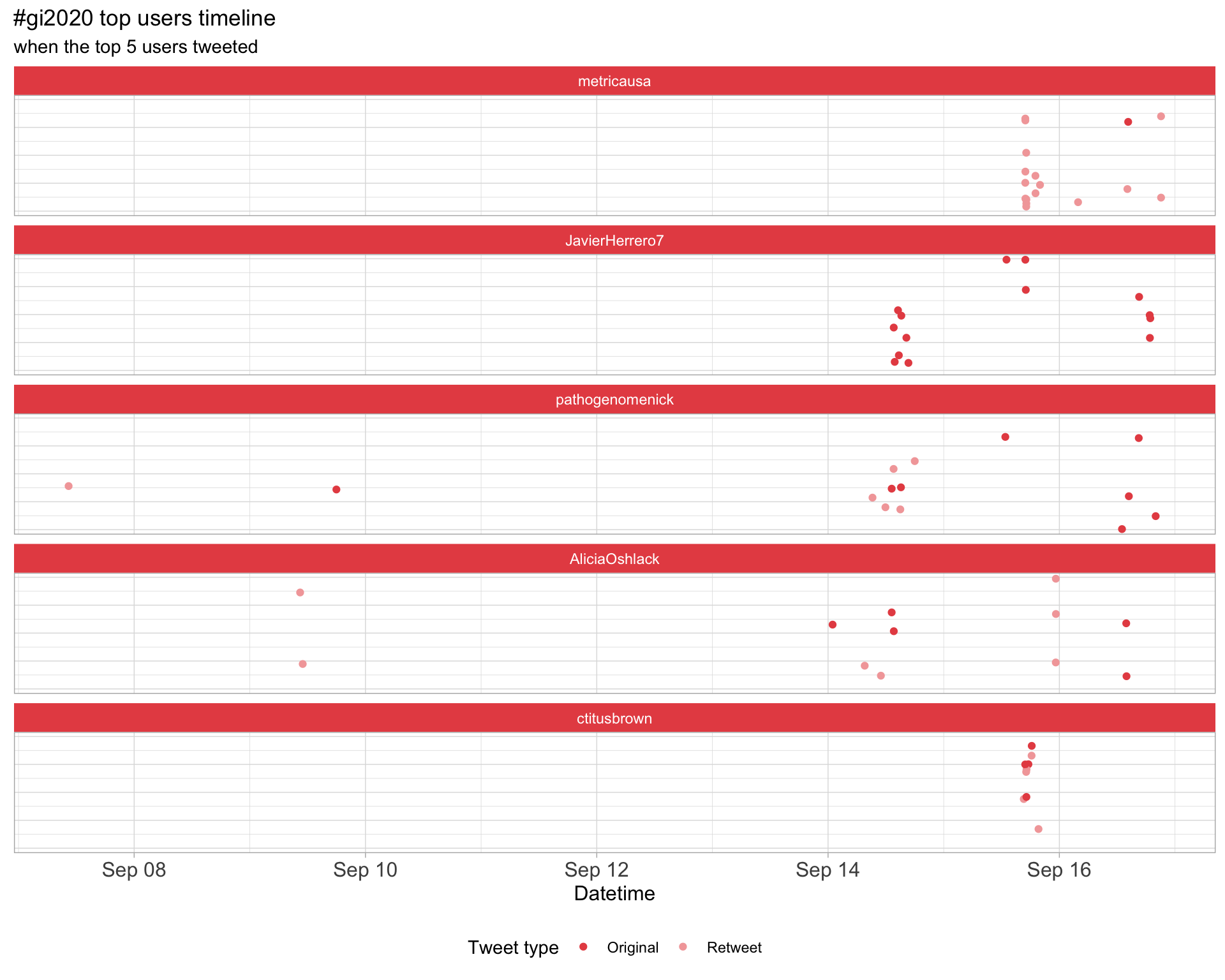
2.3 Top tweeters timeline
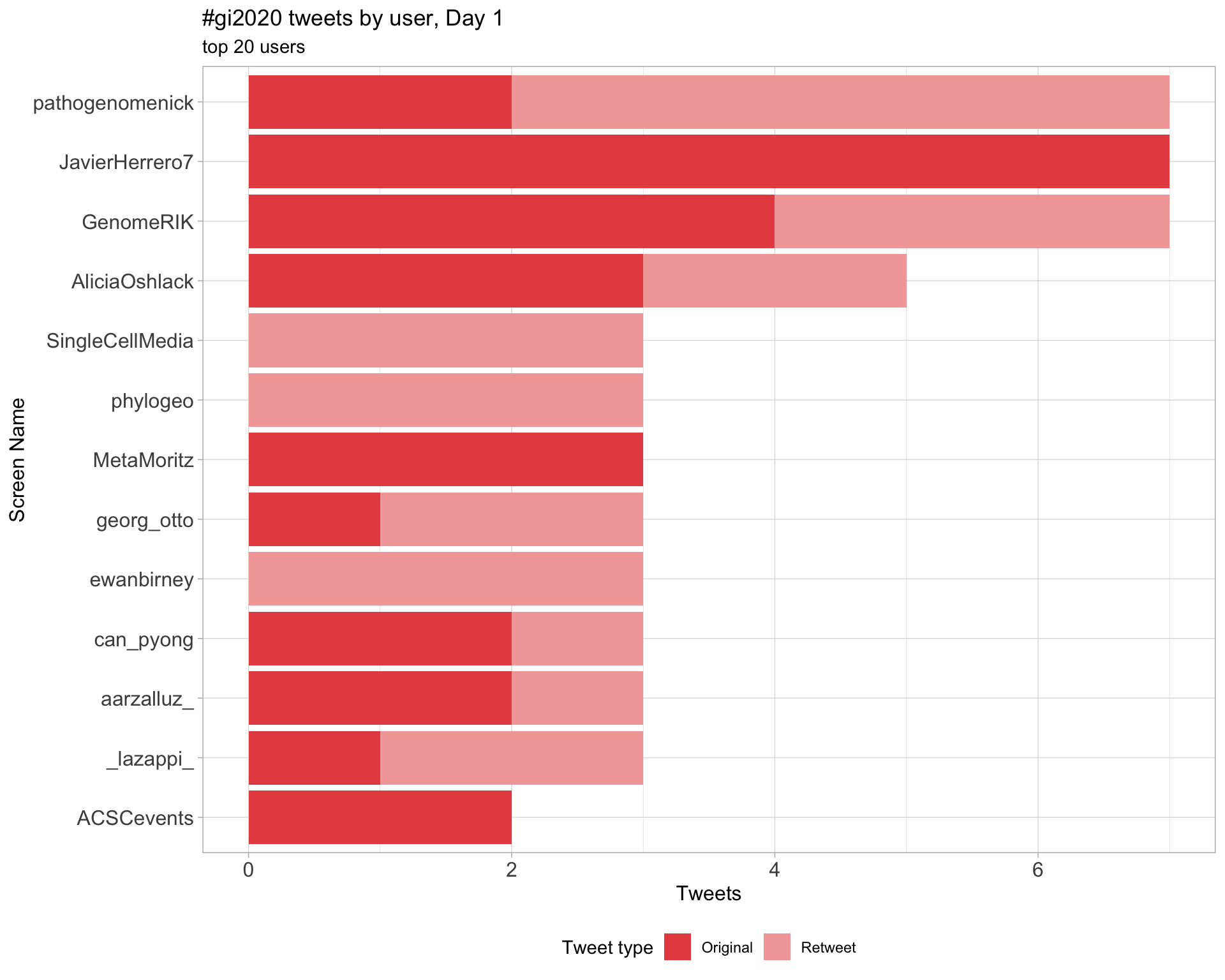
2.4 Top tweeters by day
Overall
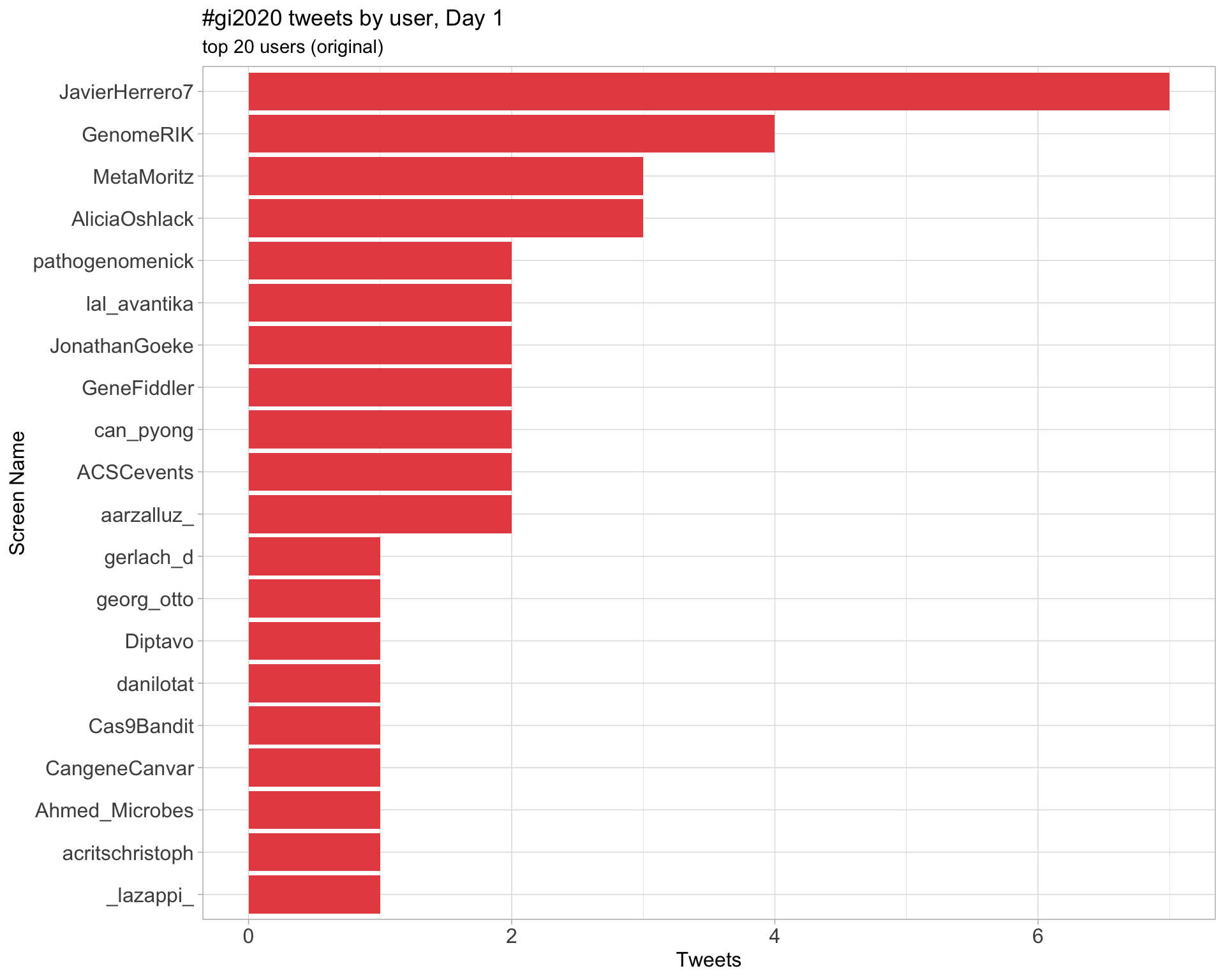
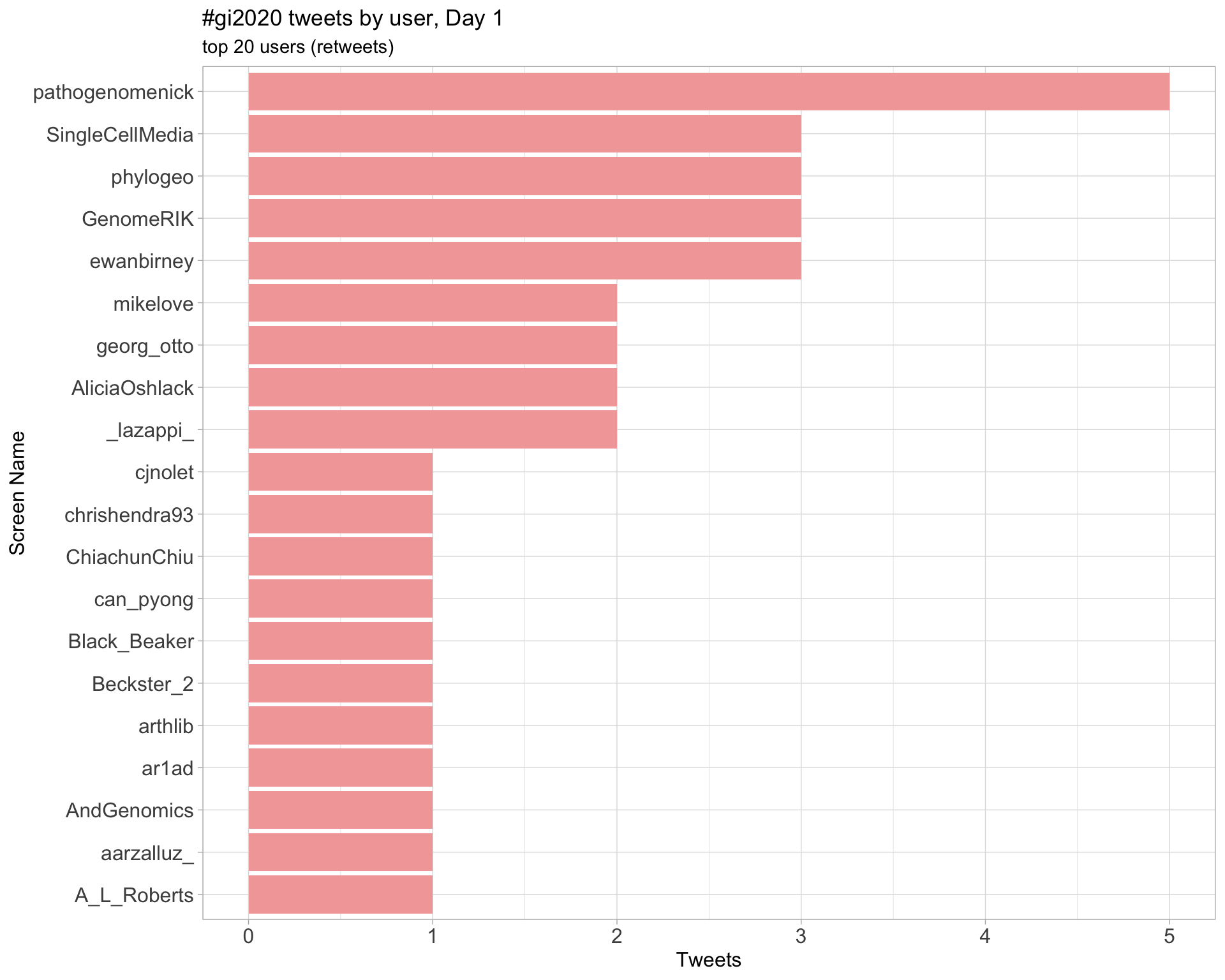
Day 1
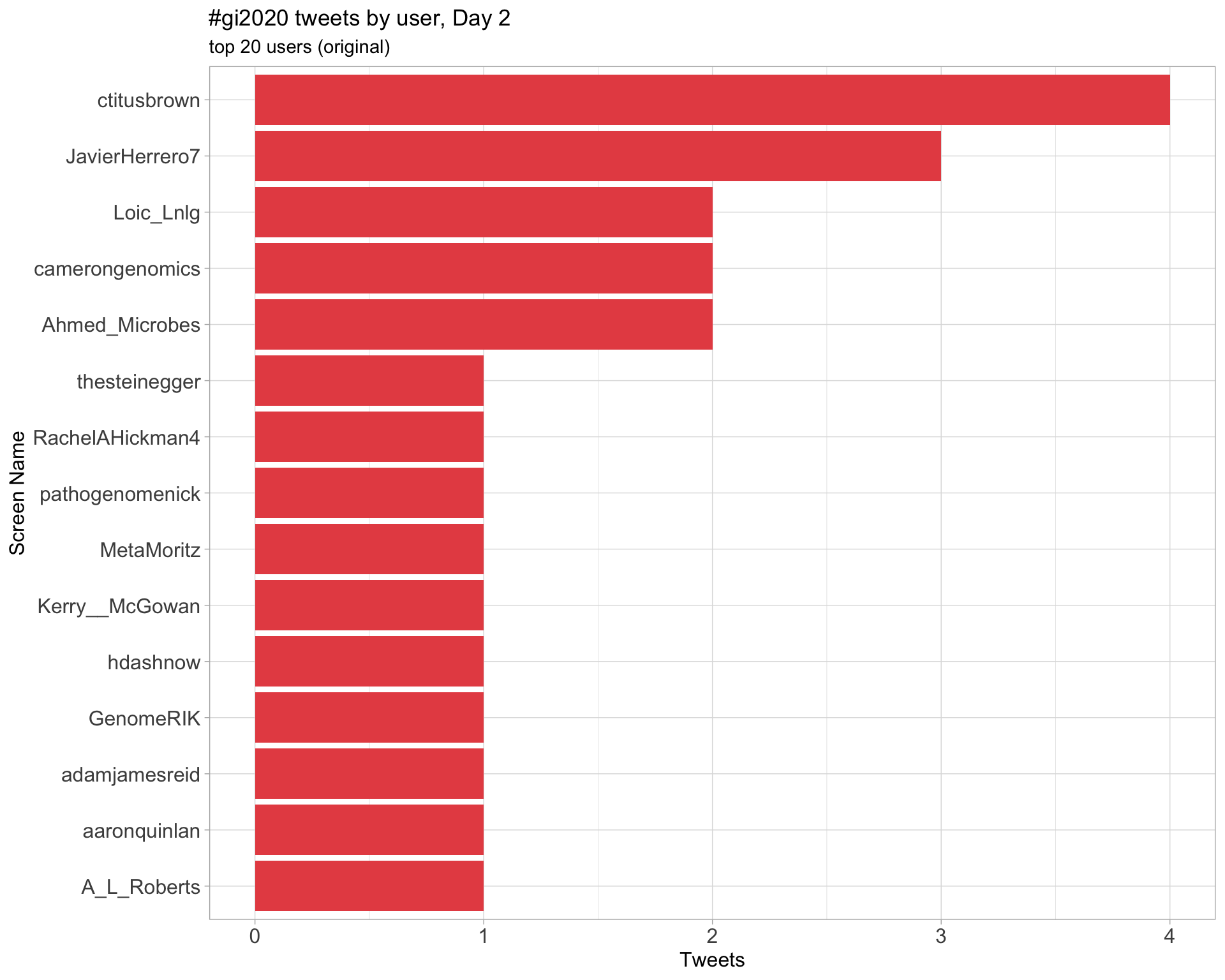
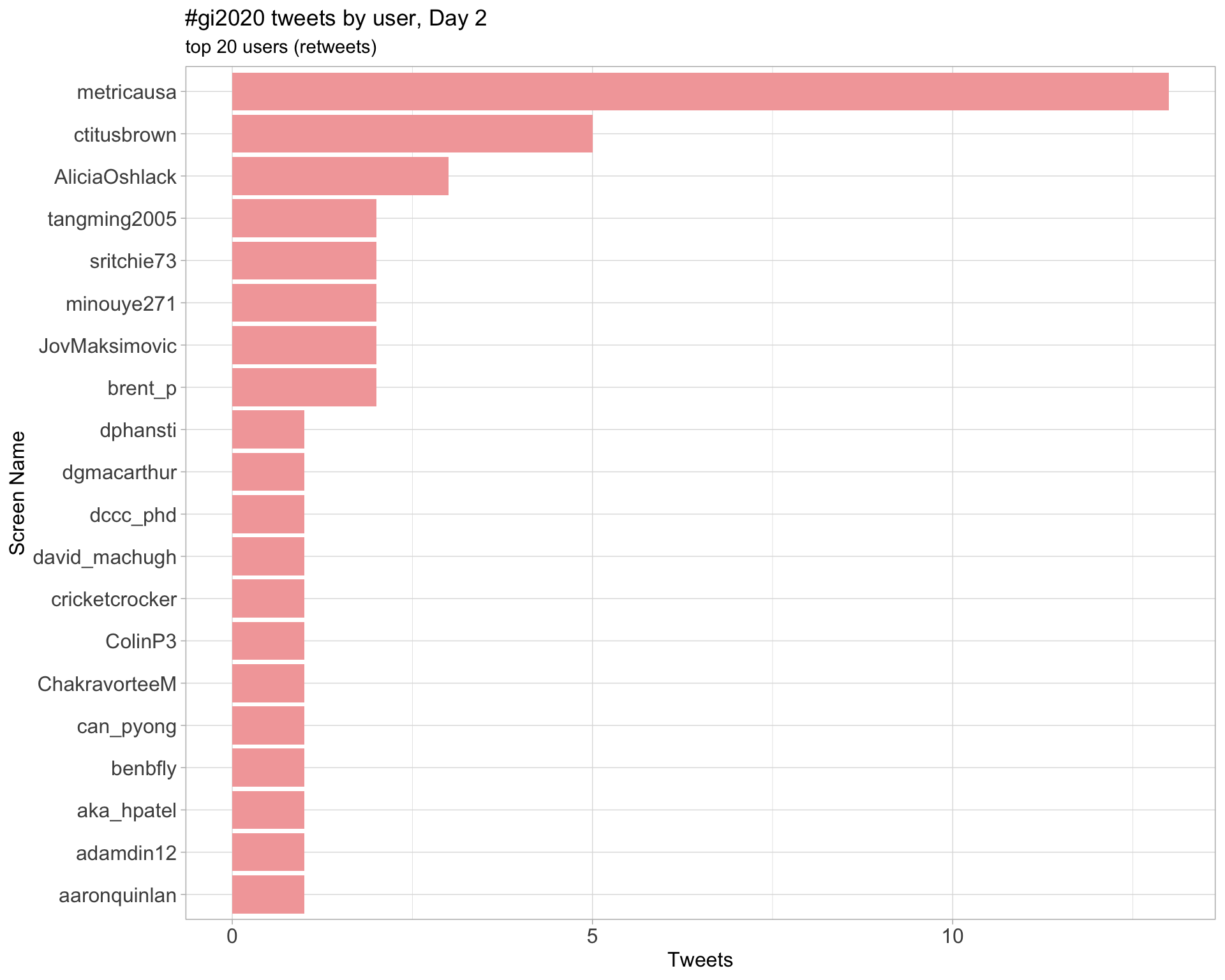
Day 2
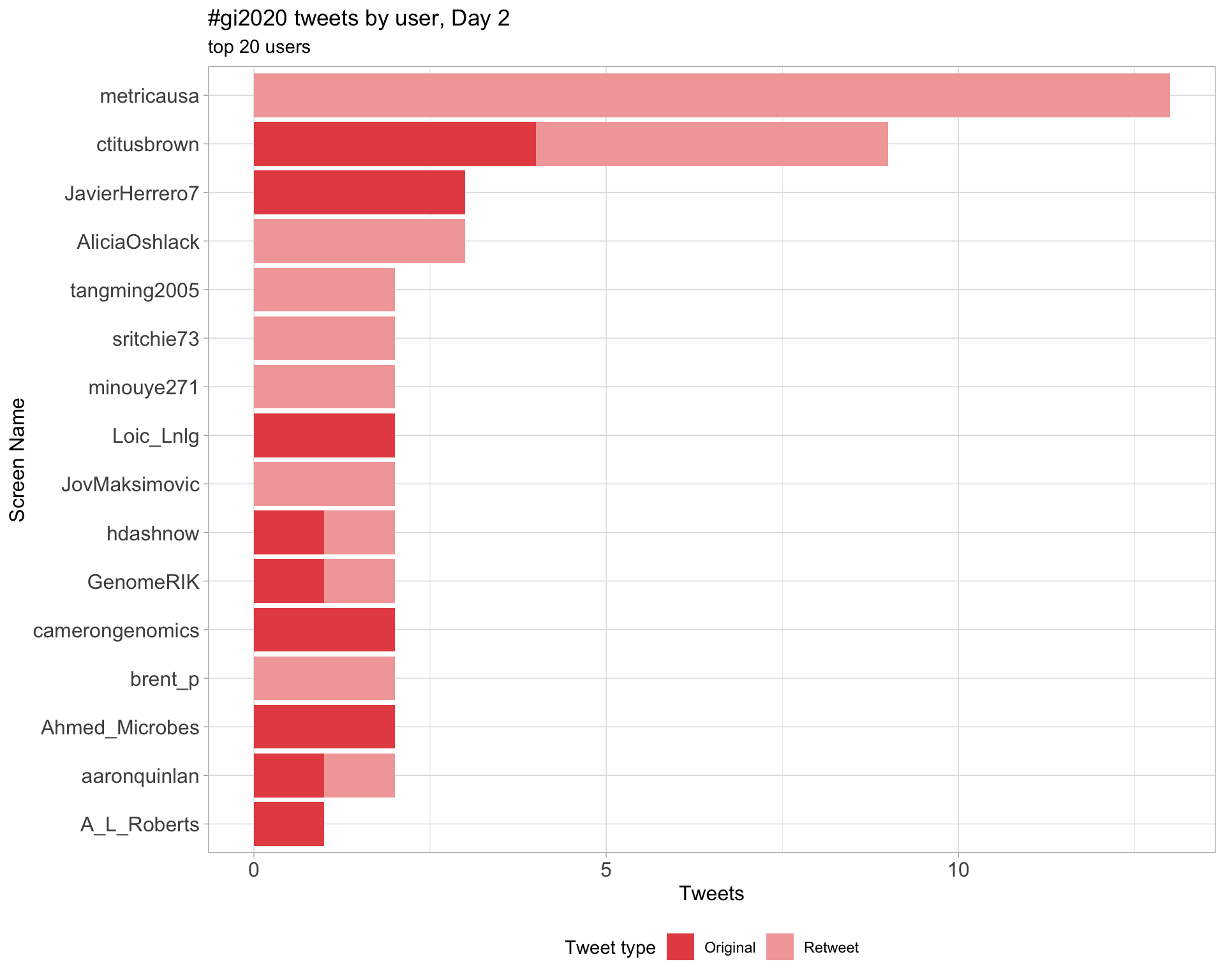
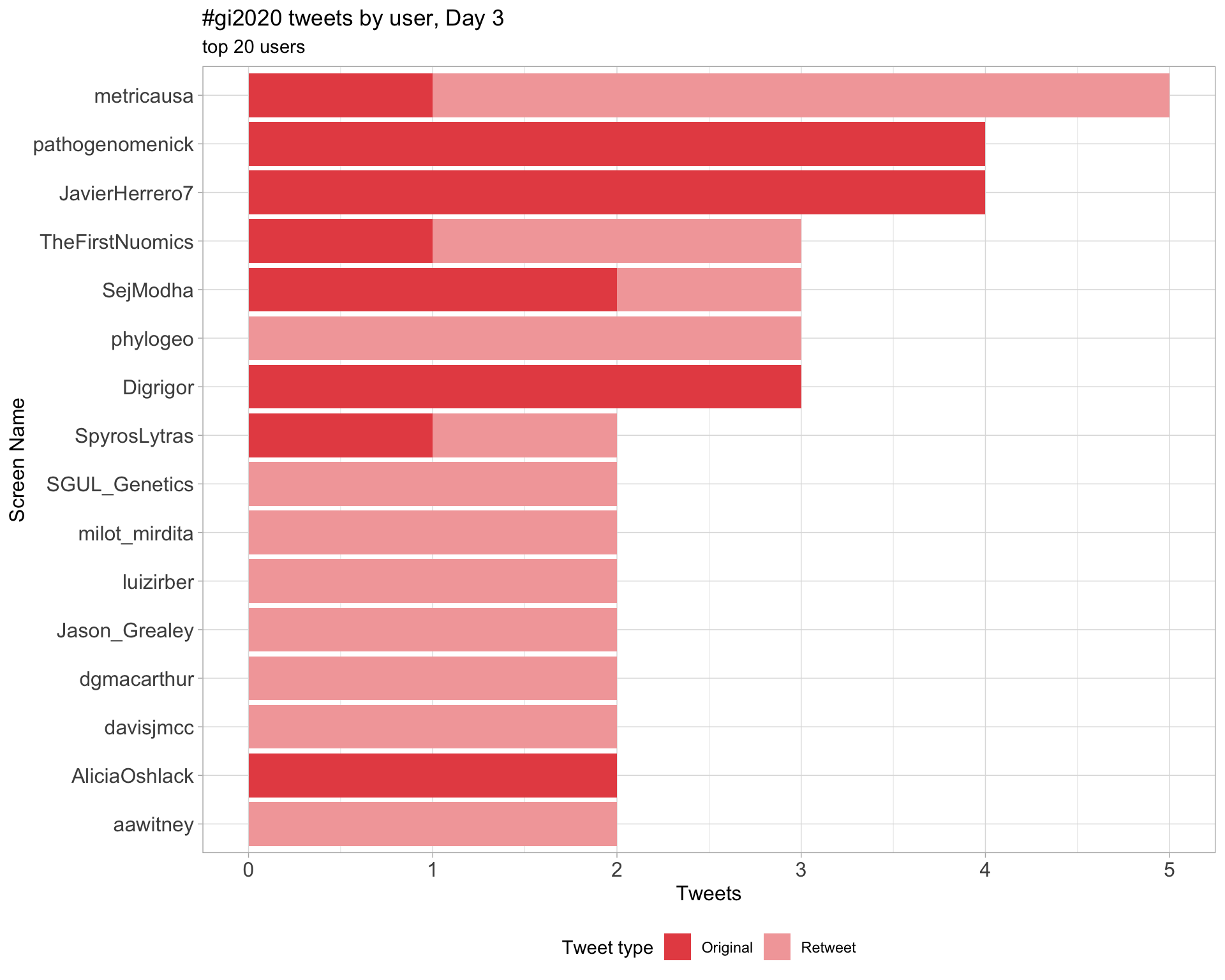
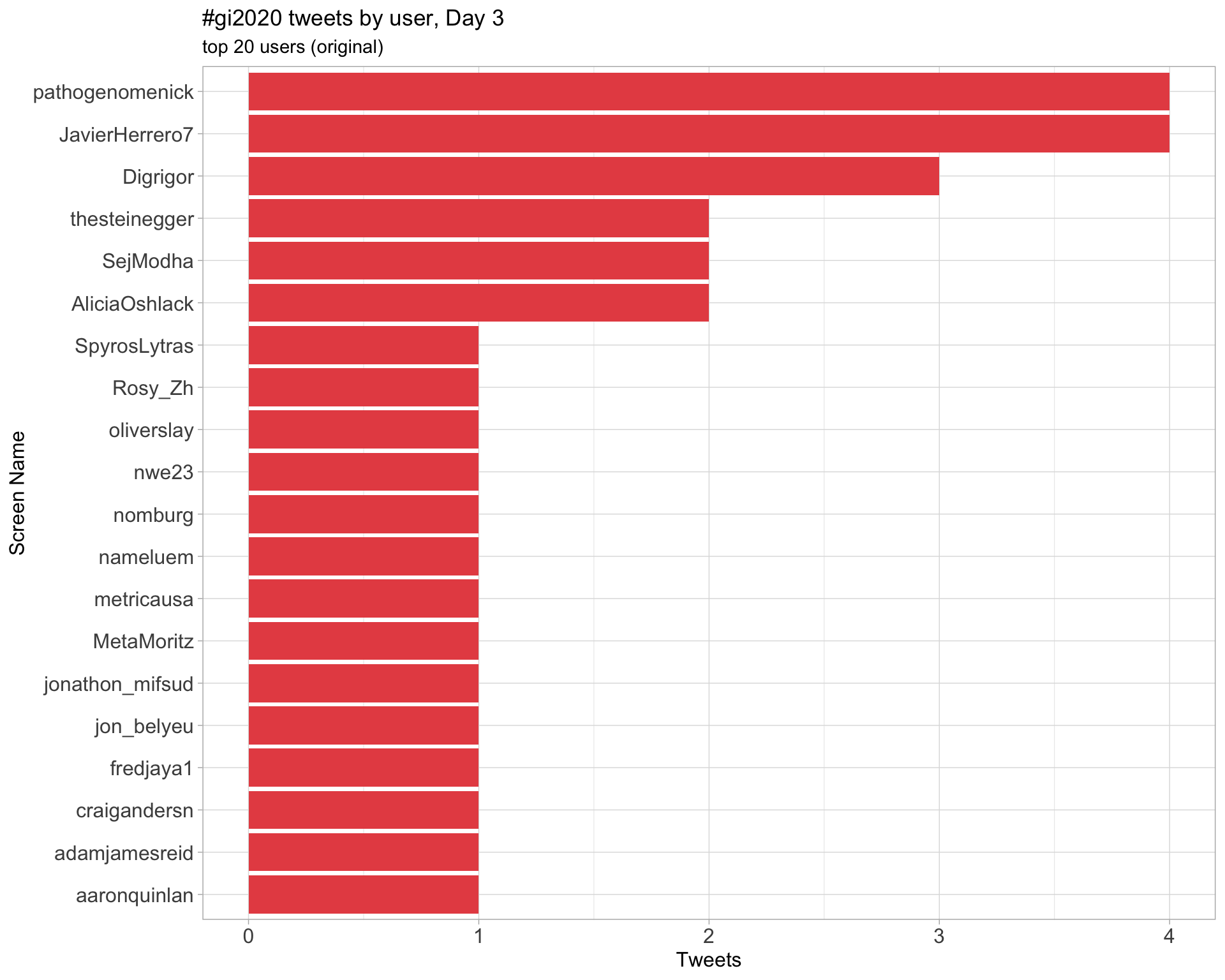
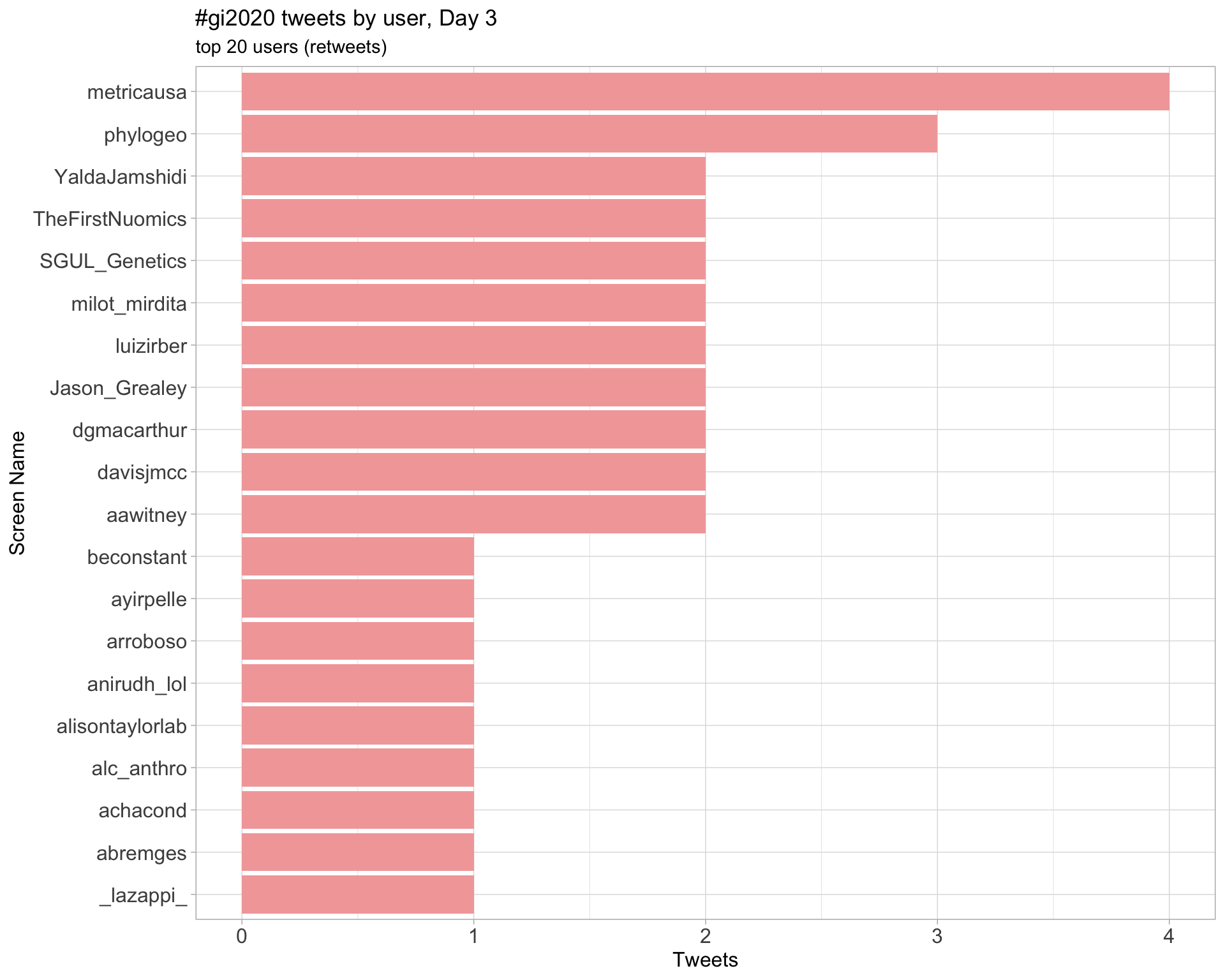
Day 3
Original
Day 1
Day 2
Day 3
Retweets
Day 1
Day 2
Day 3
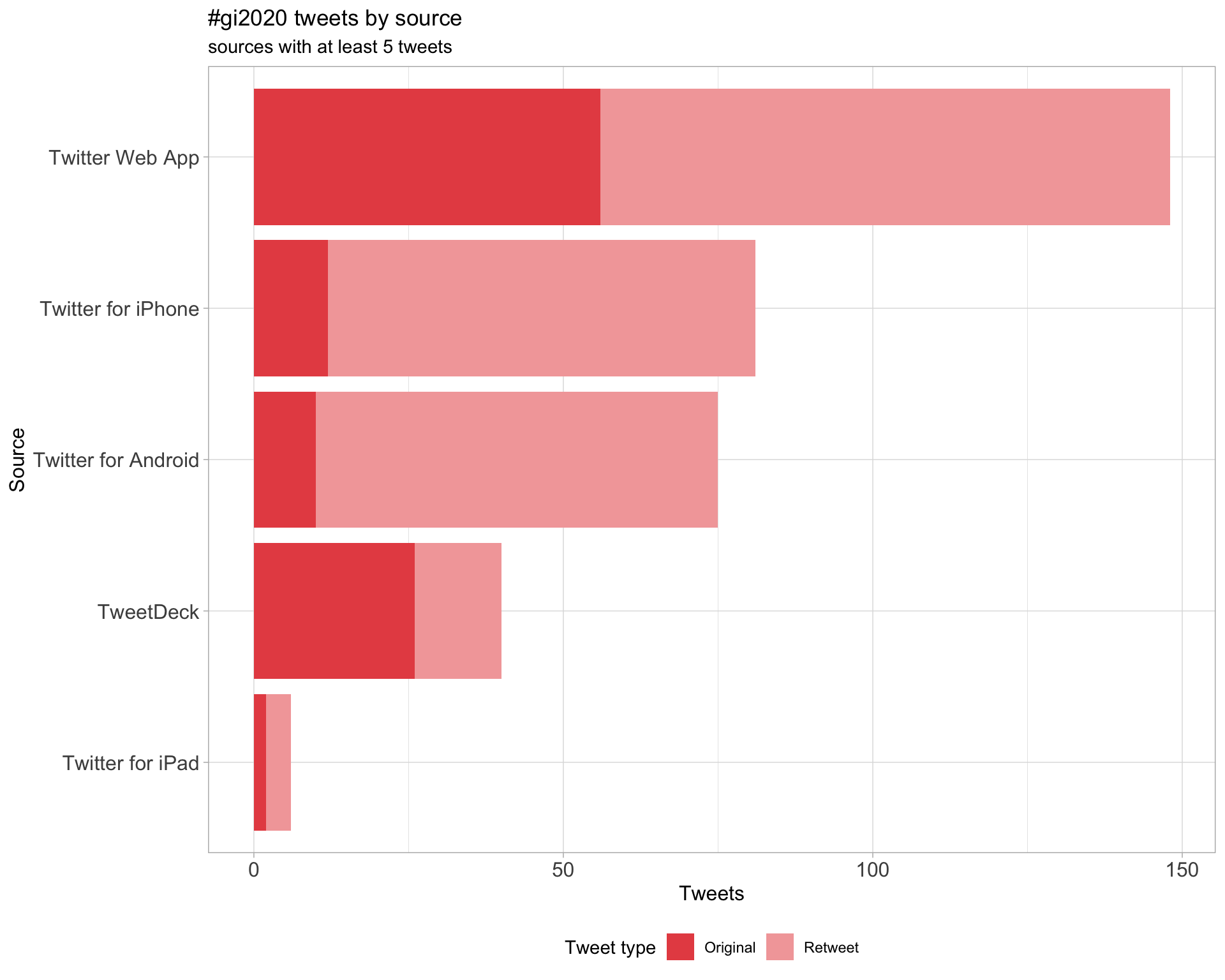
3 Sources
Users
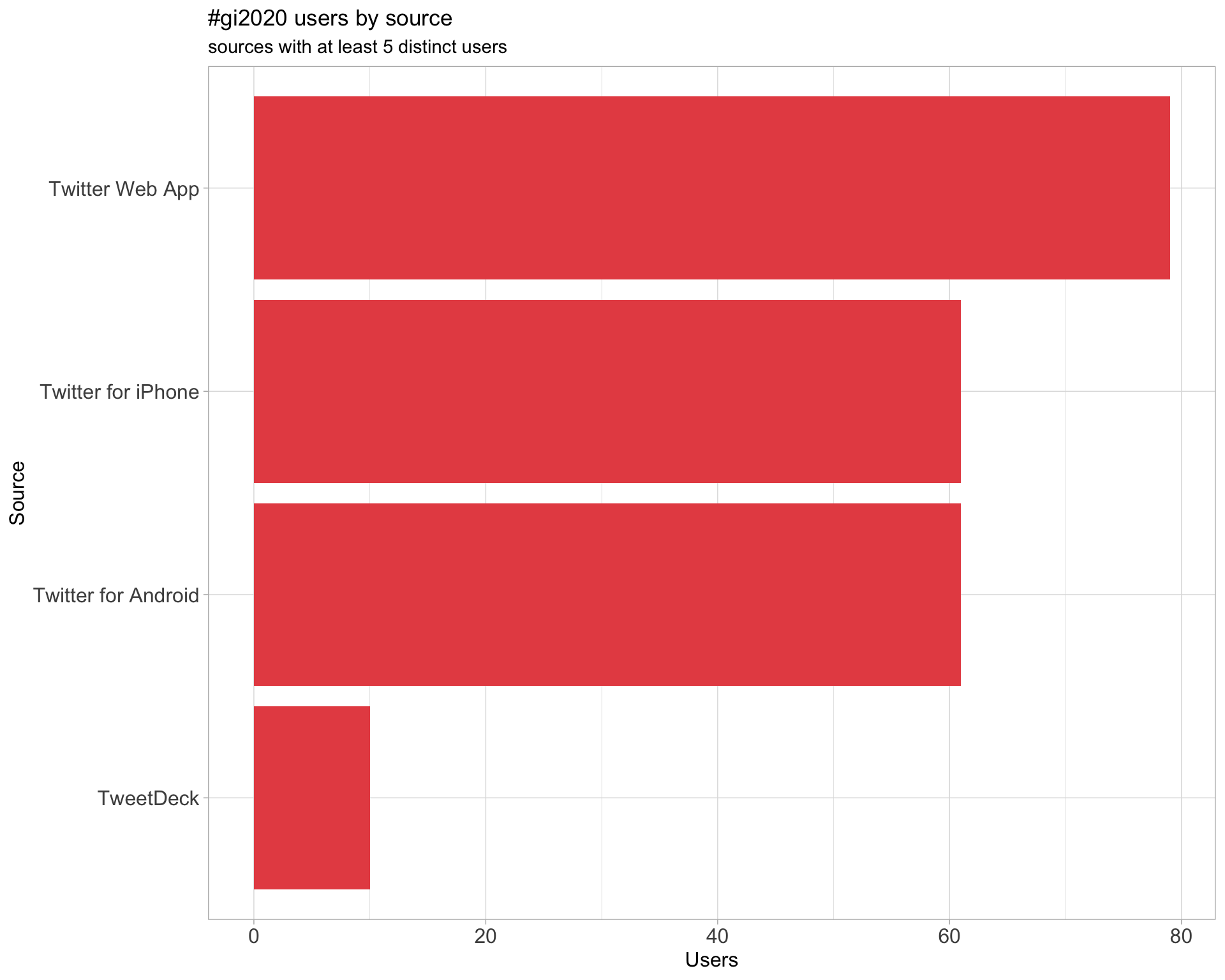
Tweets
4 Networks
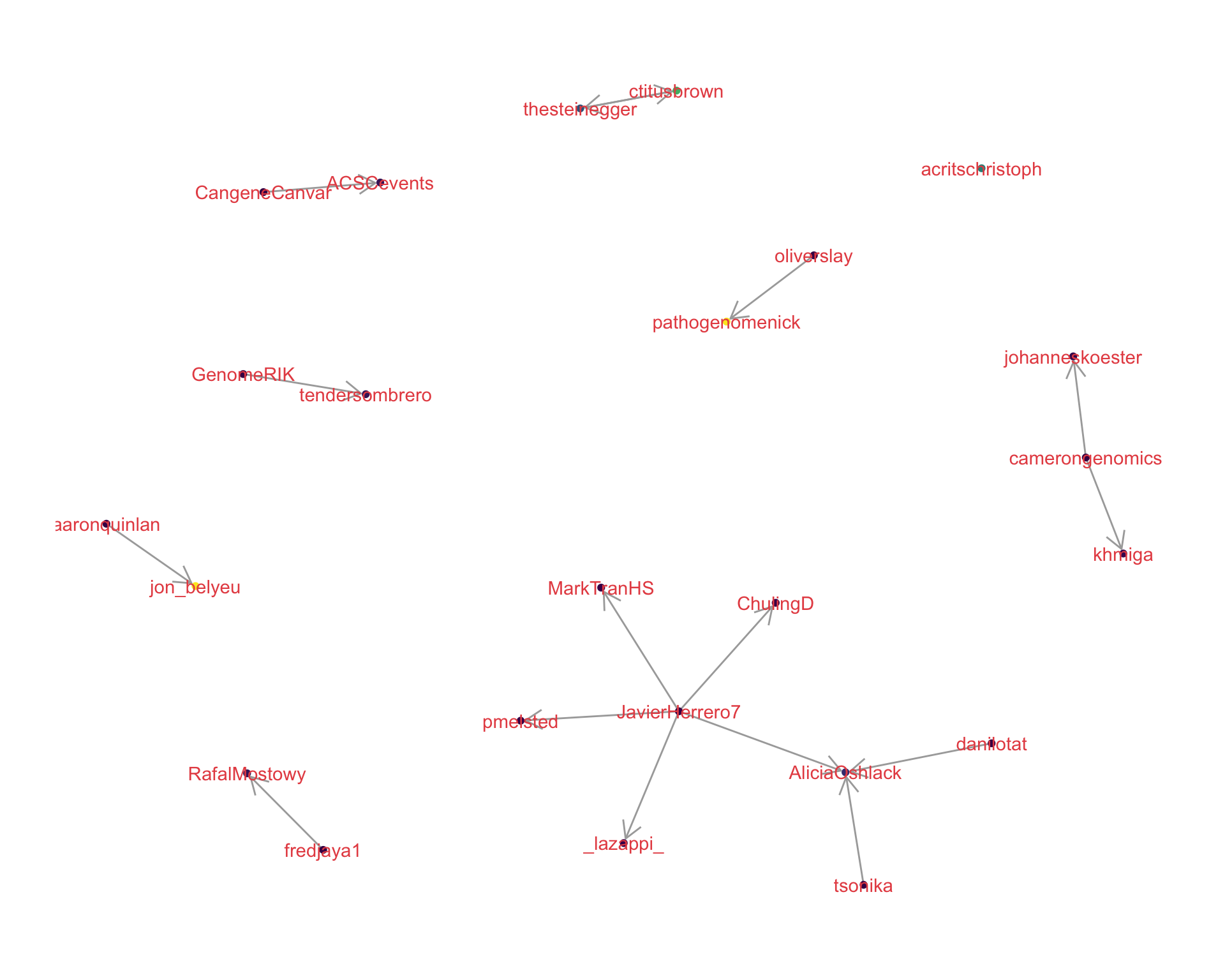
4.1 Replies
The “replies network”, composed from users who reply directly to one another, coloured by PageRank.
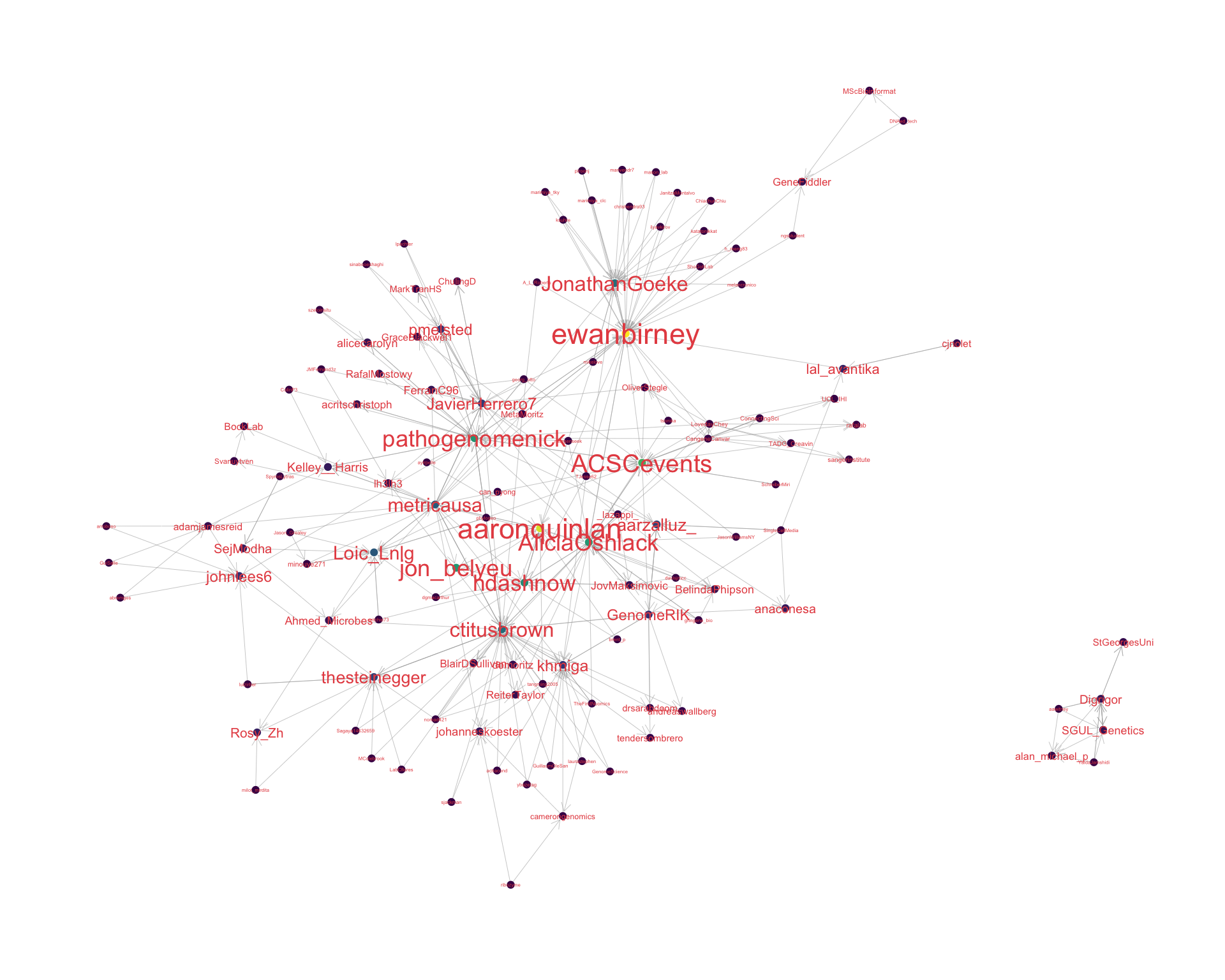
4.2 Mentions
The “mentions network”, where users mention other users in their tweets. Filtered for a k-core of 2. Node colour and size adjusted according to PageRank score.
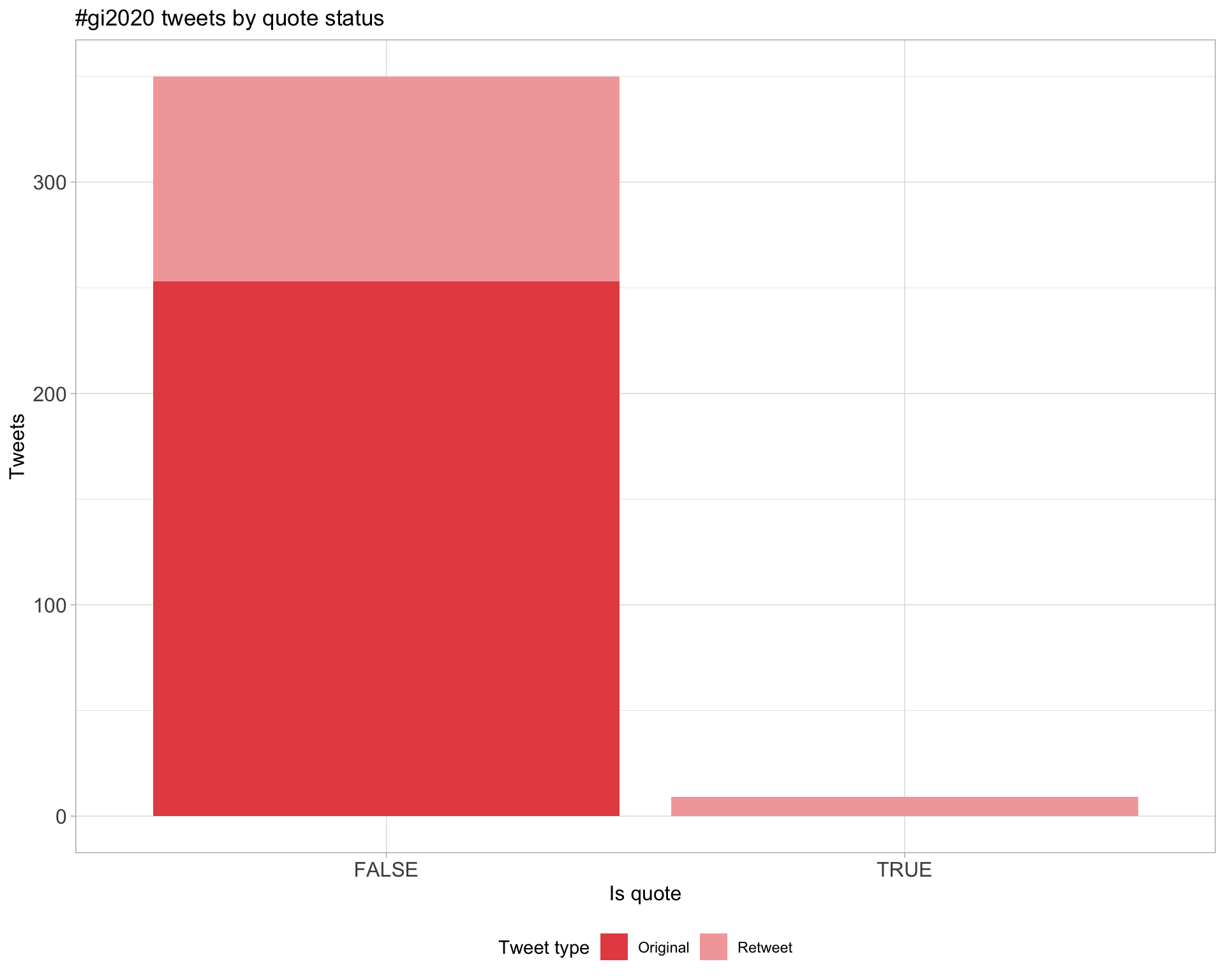
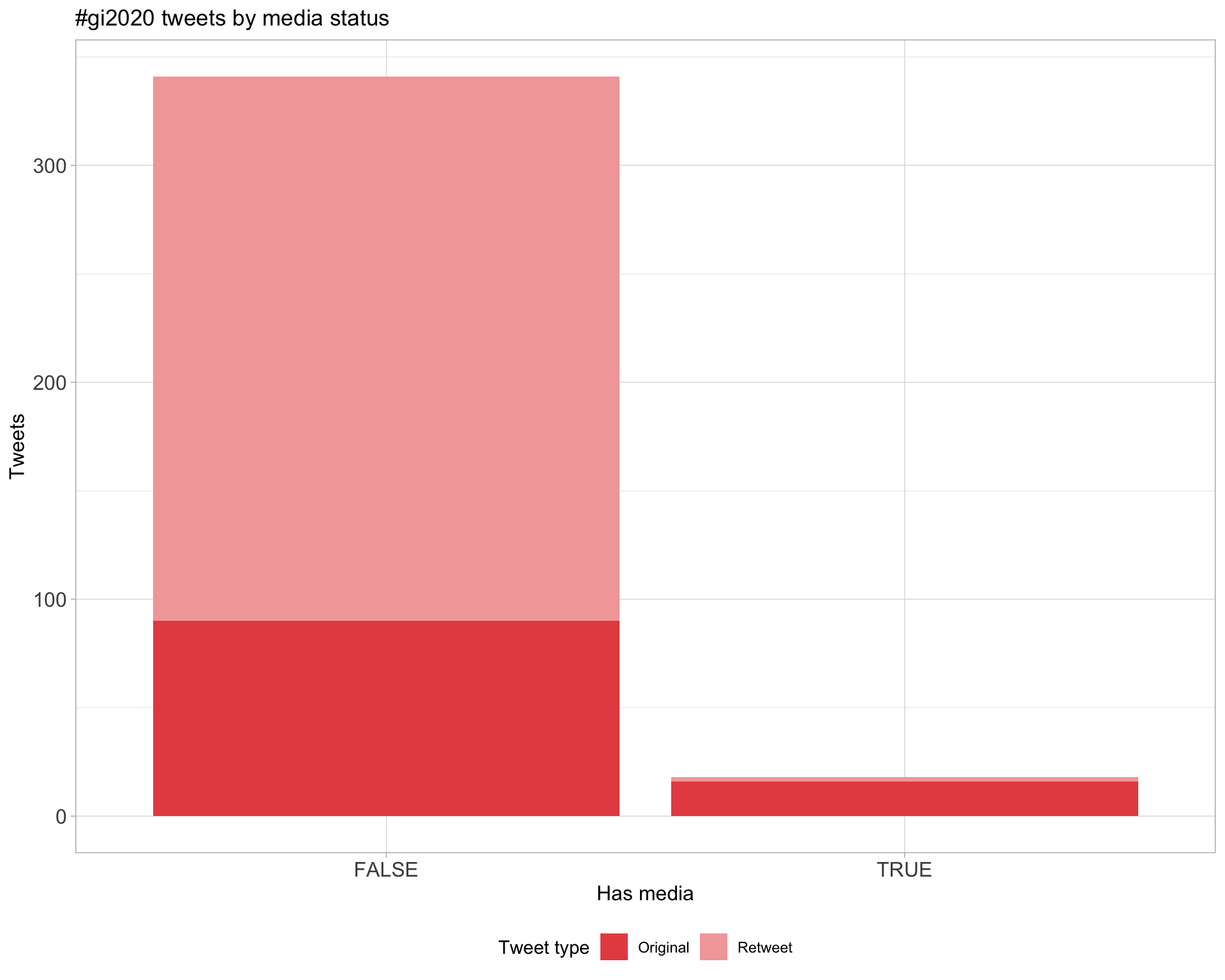
5 Tweet types
5.1 Retweets
Proportion
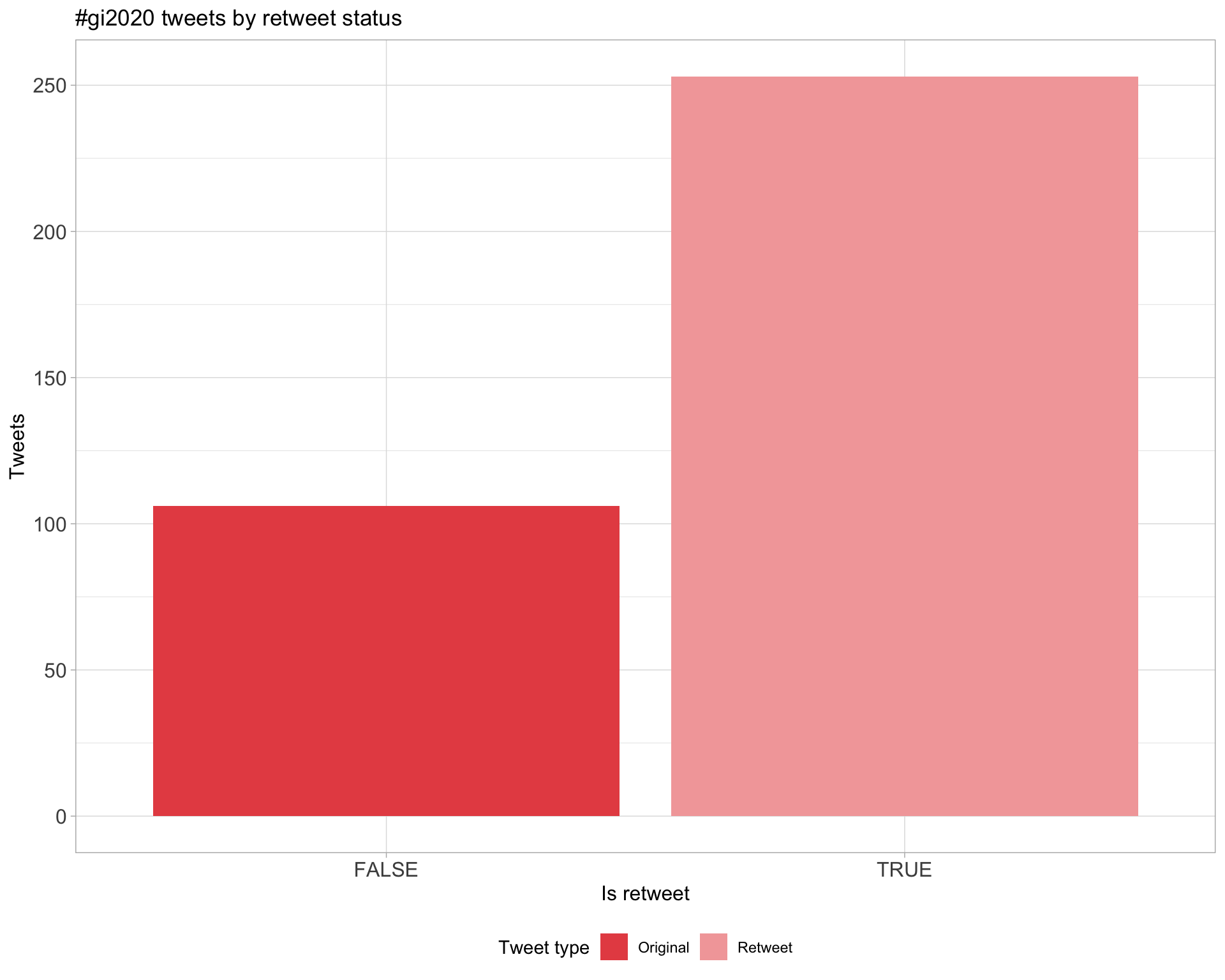
Count
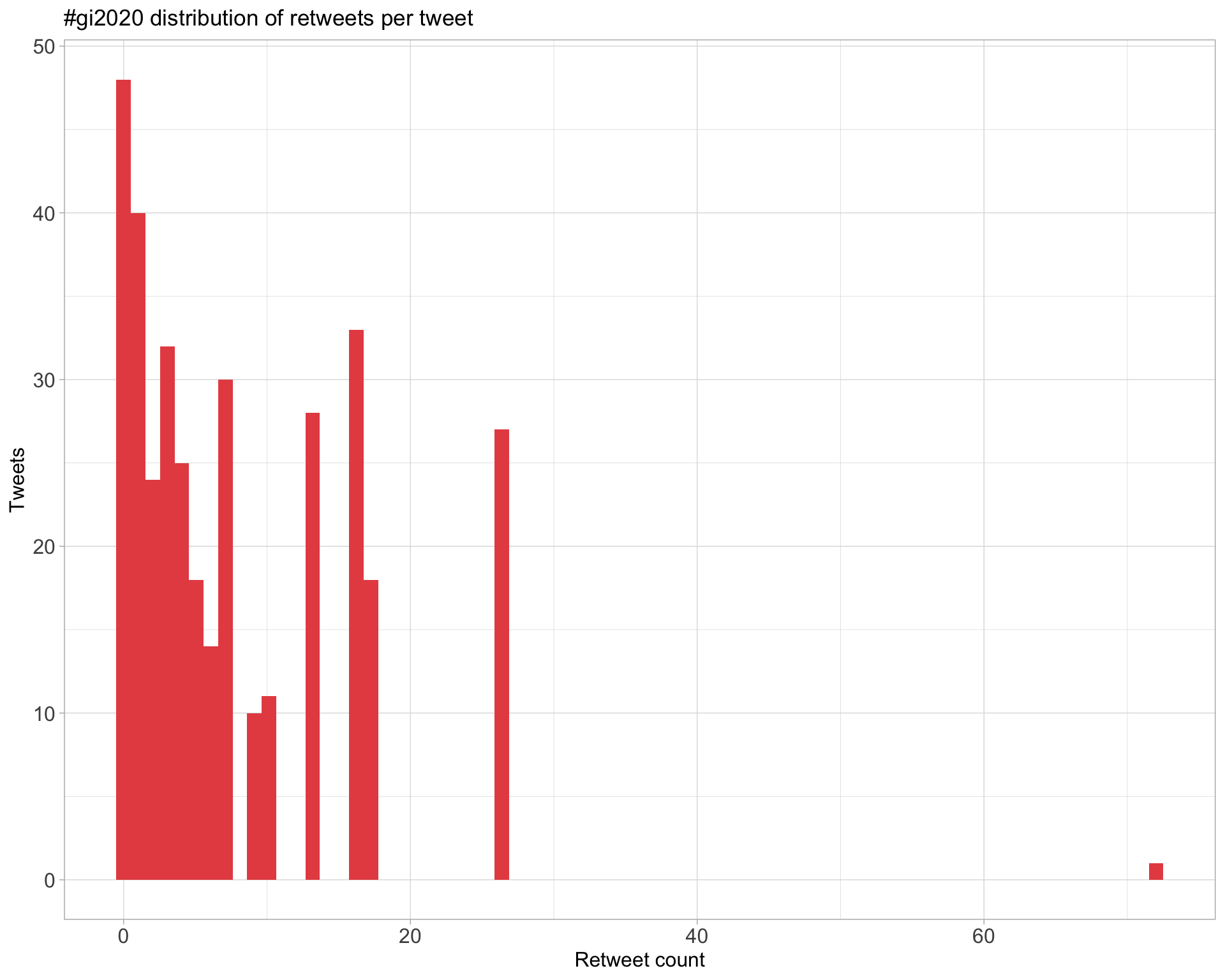
Top 10
| screen_name | text | retweet_count |
|---|---|---|
| aaronquinlan | Here is a link to our #GI2020 poster on the D4 format and associated toolset. D4 is a new format (yes I know) for representing quantitative genomics data in a more compact and faster to query manner than say, BigWig. Huge speed ups. Paper coming soon. https://t.co/YI6BmlopFZ | 26 |
| JonathanGoeke | There is a whole world of RNA modifications to be explored @ewanbirney at #GI2020. Have a look at the poster by Christopher Hendra on m6aNet to predict RNA modifications from a single sample of direct RNA-Seq, code here: https://t.co/Yuj12paUh7 | 17 |
| AliciaOshlack |
#GI2020 starts today. This is our 20th Genome Informatics meeting. What were you doing in 2001? I was working on my PhD in astrophysics on the central structure of quasars. I’m pretty sure I had no real concept of the genome 🤓 |
16 |
| ACSCevents |
#GI2020 starts today! We’re celebrating 20 years of Genome Informatics this week with over 350 participants from 42 countries. Please use the hashtag #GI2020 when discussing highlights from the meeting. Here’s a throwback of our 2001 #GenomeInformatics abstract book👇 https://t.co/NL9YQiwZ5n |
16 |
| pathogenomenick | The full Genome Informatics 2020 programme is now up, looking like a very tasty agenda indeed. About to do our technical rehearsal which I assume means loading up Zoom. #gi2020 https://t.co/yAzofXKTCH | 13 |
| Loic_Lnlg |
If you are attending Genome Informatics 2020, you can stop by our poster (P57) to learn more about the Green Algorithms project and ask questions! #GI2020 The app: https://t.co/HObEYM4fmi The preprint: https://t.co/vtqsQzh9vM https://t.co/oYlA2f3PHe |
13 |
| ctitusbrown | #GI2020 karen miga @khmiga gave great talk on sequencing, assembly, and analysis results from the telomere-to-telomere consortium! github repo here: https://t.co/C9w3bI5sPS | 10 |
| ACSCevents |
🚨FINAL CALL! Register for #GI2020 now: https://t.co/TvNUnkHKKZ Bringing researchers together to: ✅Discuss recent advances ✅Focus on approaches incl. variant-calling, #transcriptomic analysis & variant interpretation ✅Network online 📄 AGENDA: https://t.co/MMmmGEeDeM https://t.co/8Ho3nH6Hj2 |
9 |
| thesteinegger | PenguiN is a ProtEiN Guided Nucleotide assembler. It recovers many fold more viruses from metatranscriptomes than state of the art assemblers. Here is our #GI2020 Flash talk: https://t.co/I7Iawn9fMT Poster: https://t.co/WTEcdssJgq | 7 |
| hdashnow | Here’s my #GI2020 lightning talk and poster. It describes the STRling algorithm and a first look at running it on 2504 genomes from the 1000g project Talk: https://t.co/I1cYFrGHdc Poster: https://t.co/l71hgLvLzt | 7 |
Most retweeted
5.2 Likes
Proportion
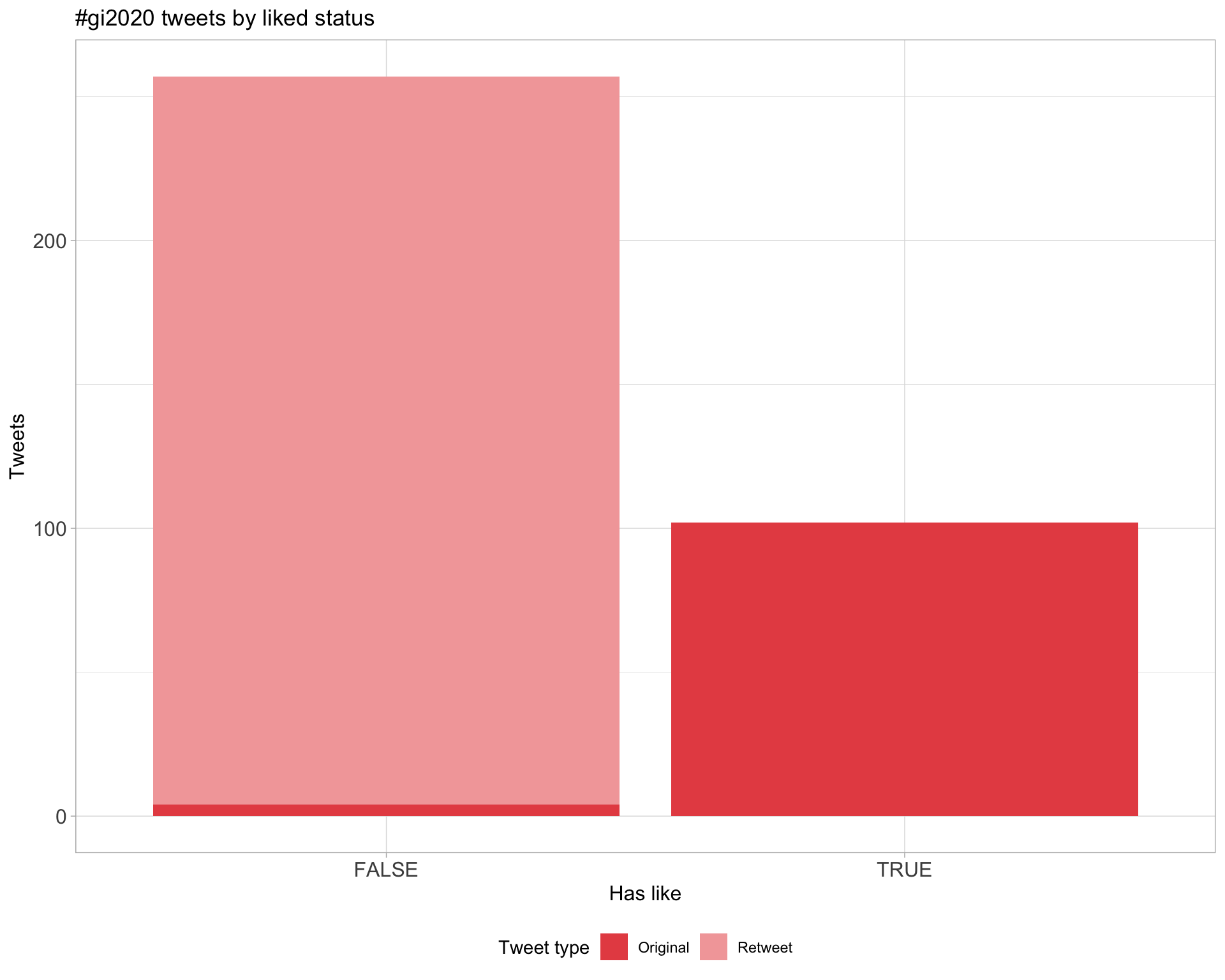
Count
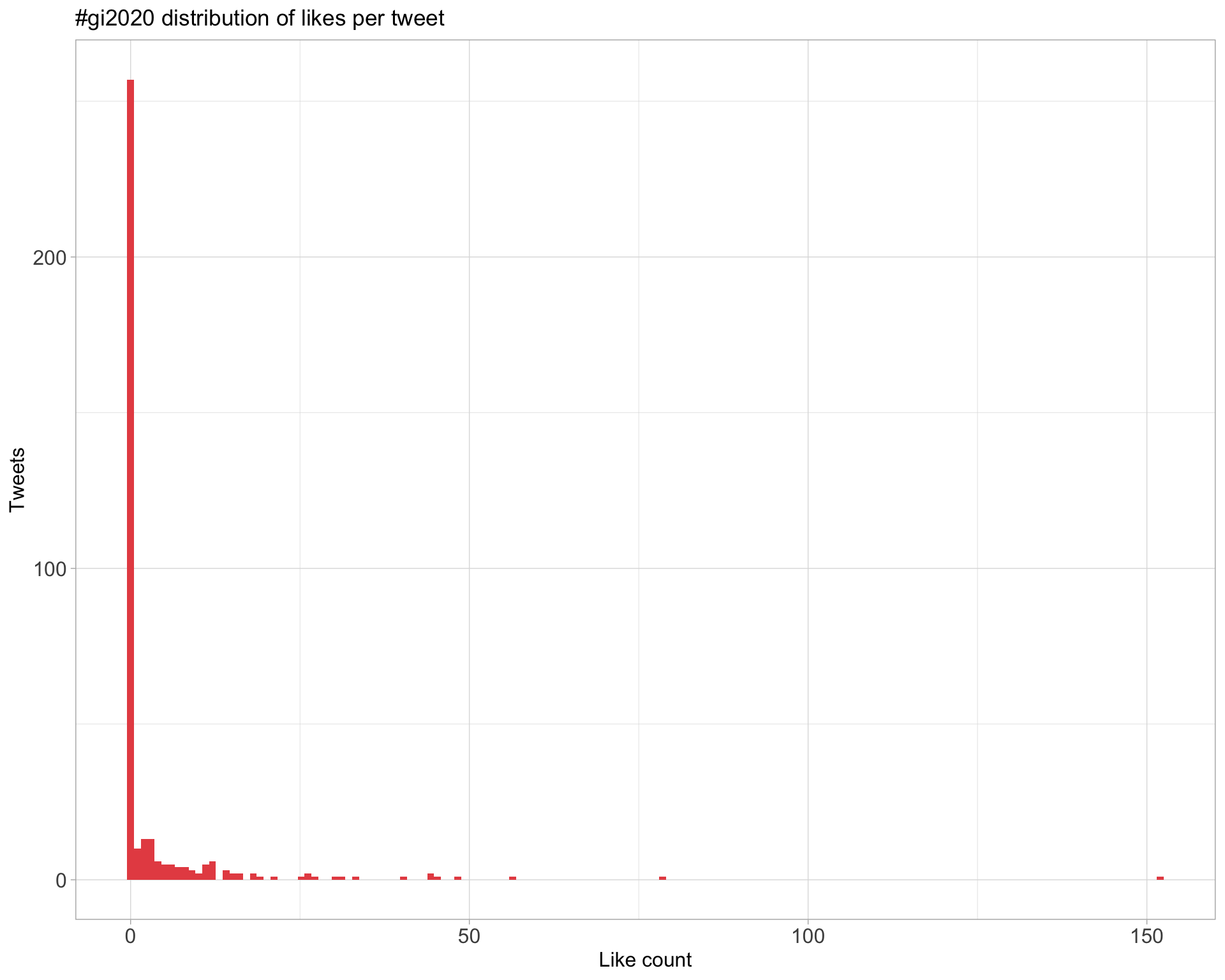
Top 10
| screen_name | text | favorite_count |
|---|---|---|
| AliciaOshlack |
#GI2020 starts today. This is our 20th Genome Informatics meeting. What were you doing in 2001? I was working on my PhD in astrophysics on the central structure of quasars. I’m pretty sure I had no real concept of the genome 🤓 |
152 |
| aaronquinlan | Here is a link to our #GI2020 poster on the D4 format and associated toolset. D4 is a new format (yes I know) for representing quantitative genomics data in a more compact and faster to query manner than say, BigWig. Huge speed ups. Paper coming soon. https://t.co/YI6BmlopFZ | 79 |
| ACSCevents |
#GI2020 starts today! We’re celebrating 20 years of Genome Informatics this week with over 350 participants from 42 countries. Please use the hashtag #GI2020 when discussing highlights from the meeting. Here’s a throwback of our 2001 #GenomeInformatics abstract book👇 https://t.co/NL9YQiwZ5n |
56 |
| pathogenomenick | Just kicking back with a beer in the Red Lion at the end of the 20th Genome Informatics, and the 1st virtual Genome Informatics #GI2020 - how was it for you? https://t.co/6tbBvWCuIV | 48 |
| JonathanGoeke | There is a whole world of RNA modifications to be explored @ewanbirney at #GI2020. Have a look at the poster by Christopher Hendra on m6aNet to predict RNA modifications from a single sample of direct RNA-Seq, code here: https://t.co/Yuj12paUh7 | 45 |
| nameluem |
New challenge (for me): present our latest paper in a one (1!) minute video.🎞️ Great format for lightning talks, as part of the 2020 (virtual) edition of the Genome Informatics meeting. #GI2020 paper: https://t.co/S5YDiJEqqz data/browser/etcetera: https://t.co/8owz00vfK1 https://t.co/1U69g8LwC9 |
44 |
| JonathanGoeke | bambu generates a curated set of transcript annotations from long read RNA-Seq data and quantifies their expression across all samples of interest. You can install bambu from github (soon on bioconductor!): https://t.co/JoemORGQoP #GI2020 | 44 |
| pathogenomenick | The full Genome Informatics 2020 programme is now up, looking like a very tasty agenda indeed. About to do our technical rehearsal which I assume means loading up Zoom. #gi2020 https://t.co/yAzofXKTCH | 40 |
| aarzalluz_ | I was 7 years old, had not even started to learn English and was nowhere near knowing what DNA was 🙈 Kind of amazing to think that today I’ll be presenting at #GI2020, first Genome Informatics meeting for me (hopefully, with many more to come). https://t.co/vD2i4cLli3 | 33 |
| pathogenomenick | Conference chairing, post-COVID edition! Listening to @ewanbirney keynote defining genome informatics. Sadly missed the photo of his early century crew cut. #GI2020 https://t.co/ZXyeDjpx9q | 31 |
Most likes
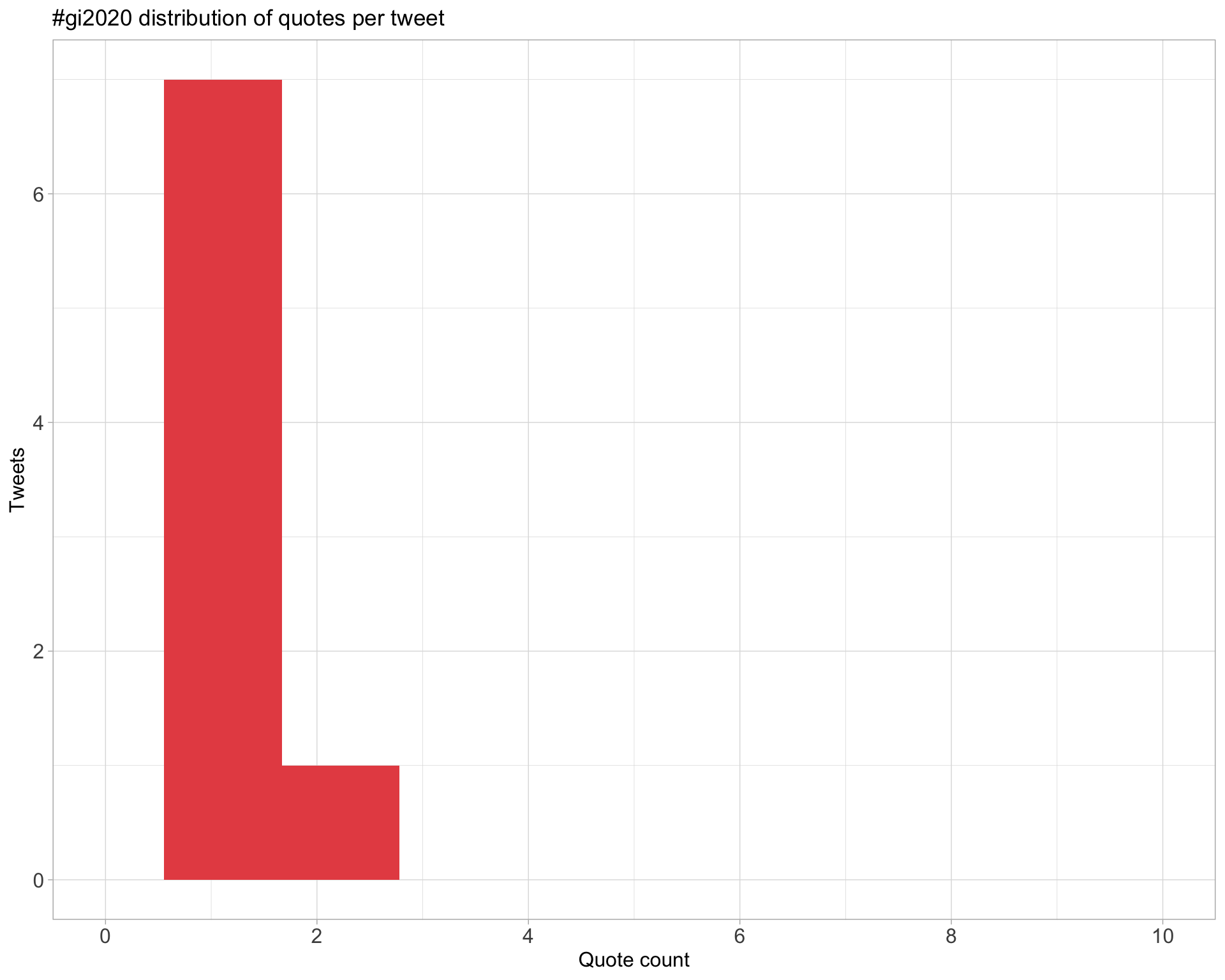
5.3 Quotes
Proportion
Count
Top 10
| screen_name | text | quote_count |
|---|---|---|
| aarzalluz_ | I was 7 years old, had not even started to learn English and was nowhere near knowing what DNA was 🙈 Kind of amazing to think that today I’ll be presenting at #GI2020, first Genome Informatics meeting for me (hopefully, with many more to come). https://t.co/vD2i4cLli3 | 2 |
| JoanaFFPViana |
In a more positive tweet, I’m really looking forward to #GI2020. The programme looks so good! PS: I was 12 in 2001 😬 https://t.co/MR2F4w6d3o |
2 |
| minouye271 | Be sure to check out Youwen (Owen) Qin’s talk at #GI2020 Wed (16th) at 130pm UK. The preprint’s just gone up! 👇 https://t.co/JzvaYuLSBP | 1 |
| MScBioinformat | #GI2020 - a very useful hashtag to watch for the next 4 days #genome #informatics #bioinformatics . Thanks @GeneFiddler ! https://t.co/VZl0Q8Gnsz | 1 |
| ACSCevents | Don’t miss this! 👇 #GI2020 https://t.co/VOYkHDnNA8 | 1 |
| Loic_Lnlg |
And if you are not at #GI2020, I got you covered! You can find my 1min lightning talk here: https://t.co/5PI91II216 and the full poster here: https://t.co/j8SO3ou8oB And you can ask your questions on Twitter! https://t.co/HObEYM4fmi https://t.co/O6ccX5sdny |
1 |
| metricausa | How green is your algorithm? What’s the carbon footprint of your GWAS, RNA read alignment, protein docking and any other computational analysis? Loïc Lannelongue @Cambridge_Uni and colleagues have an an app for that: https://t.co/Q12aw7Dn91 #Gi2020 https://t.co/3lzx3i2mSx | 1 |
| aaronquinlan | @jon_belyeu has developed a nice new tool for phasing de novo variants to gamete of origin. It uses “extended” read-backed phasing to increase the yield. #GI2020 https://t.co/UfDXo8Yp8u | 1 |
| SejModha | Really enjoyed participating in #GI2020 https://t.co/aZkhmSFE4H | 1 |
Most quoted
6 Media
Proportion
Top 10
| screen_name | text | favorite_count |
|---|---|---|
| ACSCevents |
#GI2020 starts today! We’re celebrating 20 years of Genome Informatics this week with over 350 participants from 42 countries. Please use the hashtag #GI2020 when discussing highlights from the meeting. Here’s a throwback of our 2001 #GenomeInformatics abstract book👇 https://t.co/NL9YQiwZ5n |
56 |
| pathogenomenick | Just kicking back with a beer in the Red Lion at the end of the 20th Genome Informatics, and the 1st virtual Genome Informatics #GI2020 - how was it for you? https://t.co/6tbBvWCuIV | 48 |
| nameluem |
New challenge (for me): present our latest paper in a one (1!) minute video.🎞️ Great format for lightning talks, as part of the 2020 (virtual) edition of the Genome Informatics meeting. #GI2020 paper: https://t.co/S5YDiJEqqz data/browser/etcetera: https://t.co/8owz00vfK1 https://t.co/1U69g8LwC9 |
44 |
| pathogenomenick | Conference chairing, post-COVID edition! Listening to @ewanbirney keynote defining genome informatics. Sadly missed the photo of his early century crew cut. #GI2020 https://t.co/ZXyeDjpx9q | 31 |
| aarzalluz_ | Excited to present my work today at Genome Informatics #GI2020 -tune in for some single-cell isoform fun! Great line up of speakers at the transcriptomics session this afternoon 👇🏼 https://t.co/EyD72r7RbD | 27 |
| Loic_Lnlg |
If you are attending Genome Informatics 2020, you can stop by our poster (P57) to learn more about the Green Algorithms project and ask questions! #GI2020 The app: https://t.co/HObEYM4fmi The preprint: https://t.co/vtqsQzh9vM https://t.co/oYlA2f3PHe |
19 |
| thesteinegger | Great talk from @johnlees6. He presented two tools: (1) pp-sketchlib a fast genome sketching library and (2) popPUNK a tool to cluster and visualize genomes. #GI2020. Code: https://t.co/v9v9F2idIM https://t.co/iCjFiurLXs | 16 |
| pmelsted | Handy trick to remember to look into the camera while chairing #GI2020 https://t.co/g8zXT3n4aH | 16 |
| thesteinegger | @ctitusbrown is talking about their work on spacegraphcats, a graph index structures to query diversity in metagenomes at #Gi2020. Code: https://t.co/o9FyIofTHK https://t.co/8MYOOtEtsn | 12 |
| JavierHerrero7 |
@pmelsted talking about kallisto and bustools to process scRNA-seq data. Try at home with pip install kb-python (https://t.co/oceR6woGjy) Love the slides inspired by the classic Pink Floyd’s “The Dark Side of the Moon” cover #GI2020 https://t.co/5caGEBnmFw |
12 |
6.1 Most liked image

7 Tweet text
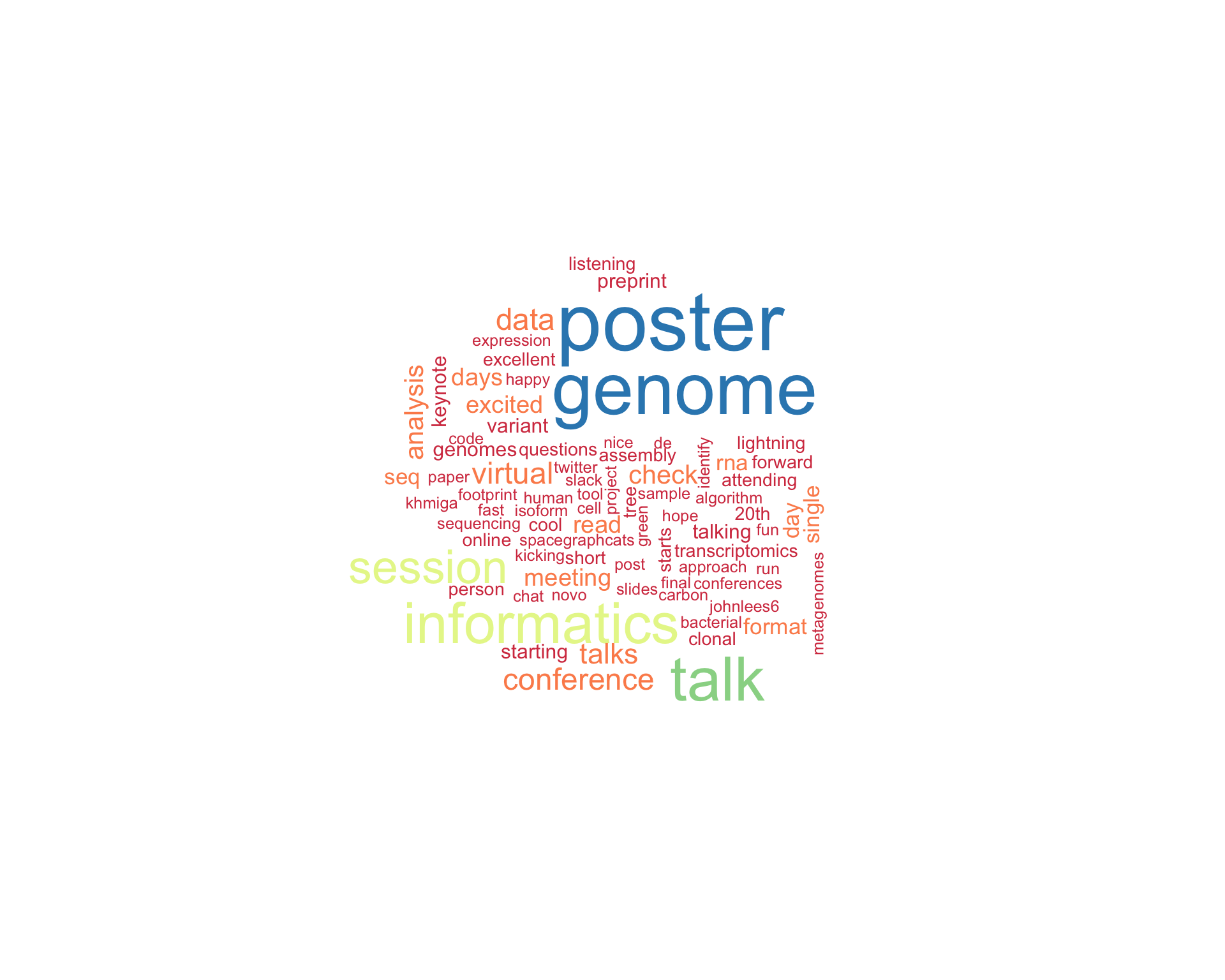
7.1 Word cloud
The top 100 words used 3 or more times.
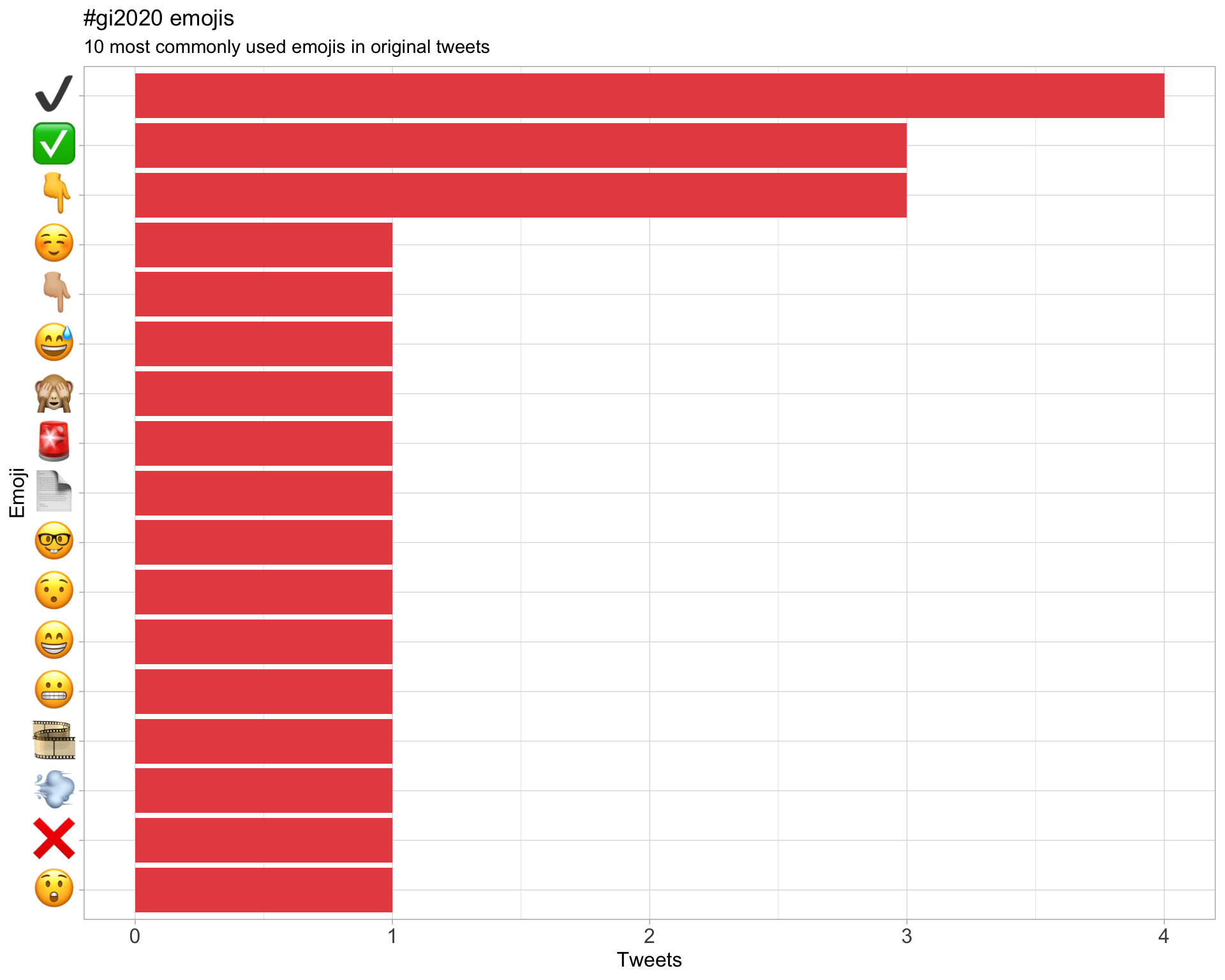
7.3 Emojis
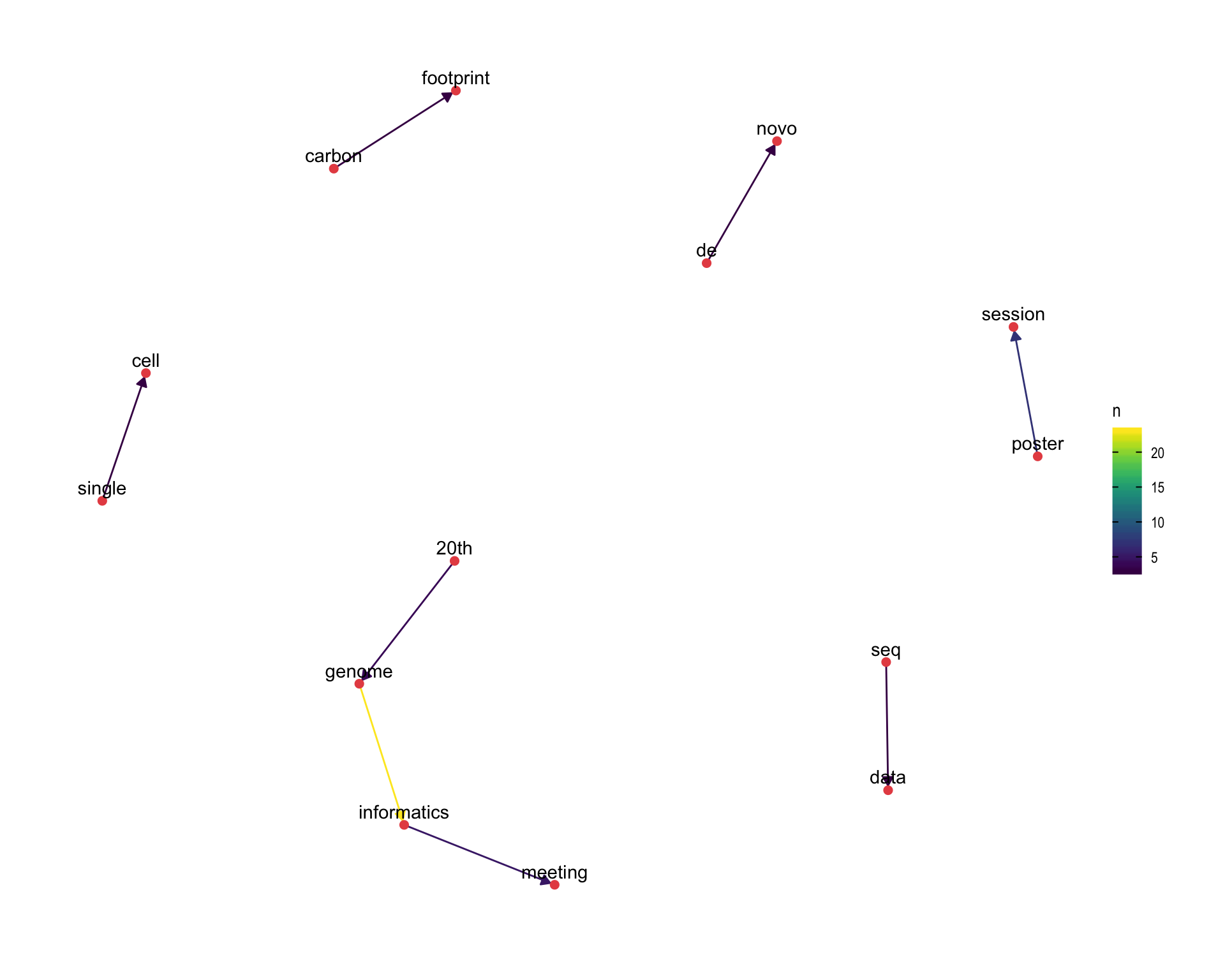
7.4 Bigram graph
Words that were tweeted next to each other at least 3 times.
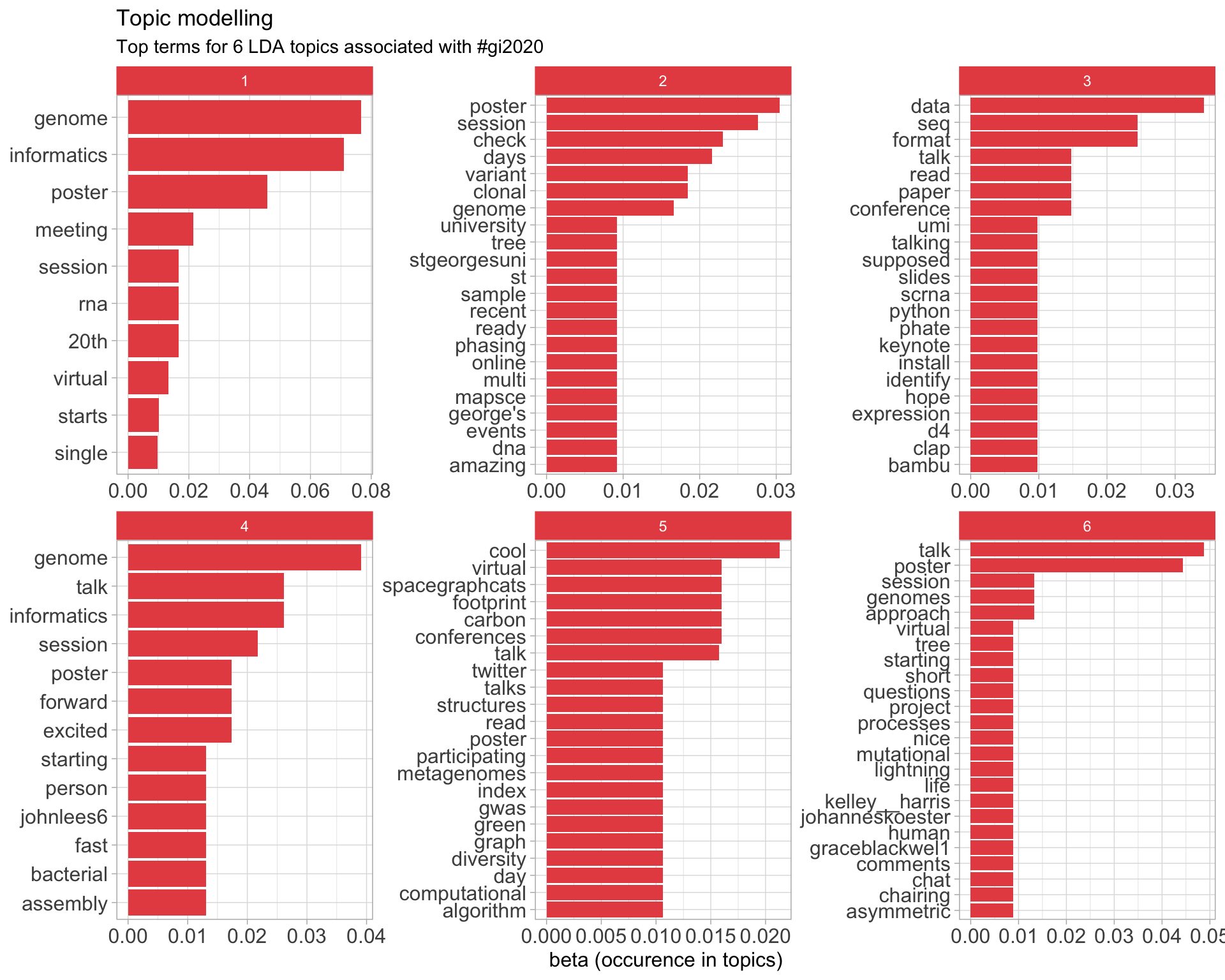
7.5 Topic modelling
Top 10 words associated with 6 topics identified by LDA.
7.5.1 Representative tweets
Most representative tweets for each topic
Topic 1
| screen_name | text | gamma |
|---|---|---|
| Diptavo | Really appreciate the immense effort the organizers have put into Genome Informatics 2020 to make it as much fun and engaging in the new virtual setup. My poster today on “Aggregative trans-eQTL associations detect trait-specific target genes in whole blood” #GI2020 https://t.co/13P7qptYOJ | 0.9953113 |
| GenomeRIK | #GI2020 #TAMAtools For anyone interested I have a poster for today’s session “P56 - Identifying novel genes in the human genome using TAMA long read RNA sequencing analysis”. It’s a bit short but I’ll be available for any TAMA questions during the poster session. | 0.9950782 |
| JonathanGoeke | There is a whole world of RNA modifications to be explored @ewanbirney at #GI2020. Have a look at the poster by Christopher Hendra on m6aNet to predict RNA modifications from a single sample of direct RNA-Seq, code here: https://t.co/Yuj12paUh7 | 0.9945349 |
| lal_avantika | After a great series of #transcriptomics and #epigenomics talks, on to the poster session at #GI2020 . #SingleCell researchers - check out @cjnolet’s poster (P80) on using @rapidsai to make single-cell analysis orders of magnitude faster! | 0.9945349 |
| GenomeRIK | #GI2020 @krsahlin What are your thoughts on NanoSplicer? Is this similar in use case as Ultra? Genome Informatics poster P127 - Accurate identification of mRNA splice sites using Oxford Nanopore sequencing | 0.9938569 |
| Loic_Lnlg |
If you are attending Genome Informatics 2020, you can stop by our poster (P57) to learn more about the Green Algorithms project and ask questions! #GI2020 The app: https://t.co/HObEYM4fmi The preprint: https://t.co/vtqsQzh9vM https://t.co/oYlA2f3PHe |
0.9938569 |
| lal_avantika | Excellent start to #GI2020 with @ewanbirney’s keynote followed by Smita Krishnaswamy. Now listening to my own recorded talk being played and feeling incredibly weird. Anyone else know this feeling? #VirtualConference | 0.9934505 |
| AliciaOshlack |
#GI2020 starts today. This is our 20th Genome Informatics meeting. What were you doing in 2001? I was working on my PhD in astrophysics on the central structure of quasars. I’m pretty sure I had no real concept of the genome 🤓 |
0.9934505 |
| pathogenomenick | Just kicking back with a beer in the Red Lion at the end of the 20th Genome Informatics, and the 1st virtual Genome Informatics #GI2020 - how was it for you? https://t.co/6tbBvWCuIV | 0.9918292 |
| JavierHerrero7 | Stick around for the last poster session #GI2020 and listen to my two PhD students (@MarkTranHS and @ChulingD) poster lightning talks. | 0.9910944 |
Topic 2
| screen_name | text | gamma |
|---|---|---|
| JavierHerrero7 | Almost ready for 2nd day of #GI2020: ✔️morning run ✔️London-style paella (AKA “rice with things”) for lunch ✔️laptop ready for notes/social interaction ✔️raspberry pi streaming talks on my TV ❌Solid internet connection 😯 | 0.9958945 |
| ACSCevents |
🚨FINAL CALL! Register for #GI2020 now: https://t.co/TvNUnkHKKZ Bringing researchers together to: ✅Discuss recent advances ✅Focus on approaches incl. variant-calling, #transcriptomic analysis & variant interpretation ✅Network online 📄 AGENDA: https://t.co/MMmmGEeDeM https://t.co/8Ho3nH6Hj2 |
0.9957169 |
| JavierHerrero7 | Redwood (Sequoia sempervirens) genome (50+Gbps) resolved in 3 days? Also assembly of the strawberry genome (octoploid) (Note: I am missing some details as my neighbours have called in the tree surgeons to trim their largely overgrown garden) #GI2020 | 0.9953113 |
| nomburg | Happy to be presenting my poster on non-canonical subgenomic RNAs generated by SARS-CoV-2 at #GI2020 (P81)! There’s a zoom link on the poster if you want to chat during the poster session. Also, feel free to check out the updated preprint - https://t.co/DVnkfZs0re | 0.9953113 |
| Digrigor | St George’s University at #Gi2020 Amazing work by @alan_michael_p and @SGUL_Genetics on creating the Middle Eastern Genome Exome Variant Browser Database. Come check his poster: Session 5, P91 @StGeorgesUni | 0.9945349 |
| Digrigor | St George’s University at #Gi2020 Come and check my poster on PQuery: A web application for visualisation and filtering of multi-sample DNA variant datasets! Session 3, P45 @StGeorgesUni @SGUL_Genetics | 0.9945349 |
| JavierHerrero7 | @MarkTranHS (P108) will be presenting MAPSCE. Following on our previous work in LOHHLA (https://t.co/V9pdRG5rLS), MAPSCE includes new features to improve mapping sub-clonal events on a tumour clonal tree. #GI2020 | 0.9938569 |
| JavierHerrero7 | @ChulingD (P31) will be presenting our recent effort to combine multi-sample EPIC data to identify clonal and sub-clonal CN events and derive all sub-clones. #GI2020 | 0.9938569 |
| aaronquinlan | @jon_belyeu has developed a nice new tool for phasing de novo variants to gamete of origin. It uses “extended” read-backed phasing to increase the yield. #GI2020 https://t.co/UfDXo8Yp8u | 0.9934505 |
| minouye271 | Be sure to check out Youwen (Owen) Qin’s talk at #GI2020 Wed (16th) at 130pm UK. The preprint’s just gone up! 👇 https://t.co/JzvaYuLSBP | 0.9910944 |
Topic 3
| screen_name | text | gamma |
|---|---|---|
| JavierHerrero7 |
@pmelsted talking about kallisto and bustools to process scRNA-seq data. Try at home with pip install kb-python (https://t.co/oceR6woGjy) Love the slides inspired by the classic Pink Floyd’s “The Dark Side of the Moon” cover #GI2020 https://t.co/5caGEBnmFw |
0.9957169 |
| aaronquinlan | Here is a link to our #GI2020 poster on the D4 format and associated toolset. D4 is a new format (yes I know) for representing quantitative genomics data in a more compact and faster to query manner than say, BigWig. Huge speed ups. Paper coming soon. https://t.co/YI6BmlopFZ | 0.9955233 |
| GeneFiddler |
Excellent talk by Angeles Arzalluz-Luque at #GI2020 describing isoform co-expression analysis in scRNA. Really like the idea of using long-read seq to identify isoforms and then short-read seq to quantify them. |
0.9948208 |
| JonathanGoeke | bambu generates a curated set of transcript annotations from long read RNA-Seq data and quantifies their expression across all samples of interest. You can install bambu from github (soon on bioconductor!): https://t.co/JoemORGQoP #GI2020 | 0.9945349 |
| JavierHerrero7 |
Attending my first virtual meeting today. #GI2020 started with @ewanbirney’s keynote address. Finding weird not to clap at the end of the talk… (would find even weirder to clap while seating alone at my dinning table) |
0.9938569 |
| JavierHerrero7 |
Smita Krishnaswamy talking about PHATE (https://t.co/mBPn2GNjBs), applied to COVID-19 data Incidentally, @FerranC96 presented the PHATE mss at our BLIC journal club last week. #GI2020 |
0.9934505 |
| pathogenomenick | Chair @pmelsted giving a cracking run down of the BUS file format and toolbox for UMI-seq data at #GI2020 - makes me wish I had some UMI data to play with! | 0.9934505 |
| GenomeRIK | @tendersombrero @drsarahdoom @andreaswallberg Just watched @khmiga talk at #GI2020 and there was a good slide showing the complementation of hifi with ont. I believe it made a good point for doing both. Sorry I think we aren’t supposed to tweet slides images for the conference. | 0.9934505 |
| AliciaOshlack | Hope you all enjoy the final day of #GI2020. It has been a lot of fun organising this conference over the last 3 years and I hope to return as a participant in future years. | 0.9918292 |
| MetaMoritz | Hanging-out awkwardly at conference coffee break where you know no one has become way less awkward at an online event! I am curious how this will work #GI2020 #geeksinthereelement #covidlife | 0.9910944 |
Topic 4
| screen_name | text | gamma |
|---|---|---|
| Rosy_Zh | Happy to present a poster at my very first conference #GI2020, on de novo prediction of phage-host relationships via CRISPR spacers using our tool SpacePHARER. Check out our poster: https://t.co/2DJgsWbkFG and the preprint: https://t.co/QGGp1dGHHa | 0.9945349 |
| thesteinegger | Great talk from @johnlees6. He presented two tools: (1) pp-sketchlib a fast genome sketching library and (2) popPUNK a tool to cluster and visualize genomes. #GI2020. Code: https://t.co/v9v9F2idIM https://t.co/iCjFiurLXs | 0.9942157 |
| JavierHerrero7 | Currently, Haoyu Cheng from @lh3lh3’s lab is talking about an de novo assembly using phased assembly graphs. GH: https://t.co/CfKno34gEz arXiv: https://t.co/rhmnbXqUG8 #GI2020 | 0.9934505 |
| jonathon_mifsud | Excited to present my poster (P73) tonight at Genome Infomatics #GI2020 - explorations of the plant virosphere. Looking forward to another late night of excellent talks. https://t.co/7MvWLktMHW | 0.9934505 |
| ctitusbrown | #GI2020 karen miga @khmiga gave great talk on sequencing, assembly, and analysis results from the telomere-to-telomere consortium! github repo here: https://t.co/C9w3bI5sPS | 0.9929866 |
| GenomeRIK | #GI2020 Genome Informatics starts today! Starting it right with transcriptomics. Looking forward to @aarzalluz_ @anaconesa talk on neural single cell isoform expression! https://t.co/L0MUH2iXL6 | 0.9929866 |
| aarzalluz_ | Excited to present my work today at Genome Informatics #GI2020 -tune in for some single-cell isoform fun! Great line up of speakers at the transcriptomics session this afternoon 👇🏼 https://t.co/EyD72r7RbD | 0.9929866 |
| pathogenomenick | And now for the second part of the session: @alicecarolyn starting with the CAMI challenge, followed by @johnlees6 on fast bacterial genome sketching and then @acritschristoph on SARS-CoV-2 strains in sewage! #gi2020 | 0.9929866 |
| can_pyong | Poster session is only 30min? 😲 Ok that’s no problem I’ll ask a question in person! 💨 I hope there is any notification to a presenter if I post a question from a website. #GI2020 | 0.9924520 |
| pathogenomenick | The full Genome Informatics 2020 programme is now up, looking like a very tasty agenda indeed. About to do our technical rehearsal which I assume means loading up Zoom. #gi2020 https://t.co/yAzofXKTCH | 0.9918292 |
Topic 5
| screen_name | text | gamma |
|---|---|---|
| Ahmed_Microbes | Just got introduced to The Green Algorithm by @Loic_Lnlg at #GI2020 that quantifies the carbon footprint of genomics. The calculator (https://t.co/X928bQ4lJx) is easy to use and encourages scientists to report/watch/reduce their computational carbon footprint. #GlobalWarming | 0.9948208 |
| CangeneCanvar |
@ACSCevents @ewanbirney @AliciaOshlack @pathogenomenick @rafalab @OliverStegle @TADG_Creavin @UCL_IHI @sangerinstitute @ConnectingSci Our Higher Scientific Officer @LovedayChey has been enjoying the diverse range of talks at #GI2020 today - roll on day 2! You can read about the teams #WES and #GWAS work in #TesticularCancer in our blog: https://t.co/iny7mr9f1q https://t.co/eODY1IJXBI |
0.9948208 |
| metricausa | How green is your algorithm? What’s the carbon footprint of your GWAS, RNA read alignment, protein docking and any other computational analysis? Loïc Lannelongue @Cambridge_Uni and colleagues have an an app for that: https://t.co/Q12aw7Dn91 #Gi2020 https://t.co/3lzx3i2mSx | 0.9948208 |
| A_L_Roberts | End of Day 2 of my first virtual conference #Gi2020 - it’s been briiant so far! It’s run to time thanks to pre-recorded talks. Live Q+As at the end of each sessions where all speakers are present have been engaging. Here’s hoping virtual conferences increase inclusivity too! | 0.9945349 |
| ctitusbrown | @thesteinegger my talk slides - https://t.co/f5I0jjHztJ - work with @BlairDSullivan @domoritz @ReiterTaylor w/Michael O’Brien and Felix Reidl (whose twitter handle I can’t remember). #gi2020 | 0.9924520 |
| thesteinegger | @ctitusbrown is talking about their work on spacegraphcats, a graph index structures to query diversity in metagenomes at #Gi2020. Code: https://t.co/o9FyIofTHK https://t.co/8MYOOtEtsn | 0.9910944 |
| AliciaOshlack | Checkout the poster P90 from @BelindaPhipson and @JovMaksimovic from my group on gene set testing methods for methylation arrays #GI2020 | 0.9902143 |
| SpyrosLytras |
One of the best-organised virtual conferences I’ve been to so far! lots of new ideas and concepts at #GI2020 Many thanks to the organisers! @pathogenomenick kudos to @SejModha for her excellent poster! 😁 |
0.9902143 |
| Ahmed_Microbes | Great talk by @ctitusbrown about spacegraphcats, a graph index structures to facilitate querying diversity in metagenomes #Gi2020. | 0.9891411 |
| TheFirstNuomics | Find out more about the technology and data behind Coronavirus Network Explorer in this just released preprint #GI2020 https://t.co/GCemnxosop | 0.9878036 |
Topic 6
| screen_name | text | gamma |
|---|---|---|
| can_pyong | Thank you very much for listening my talk about a meta chromatin accessibility analysis. Although a preprint hasn’t been published yet, I’m happy to exchange the ideas with ATAC-people! I also get excited to join a poster with being free from pressure☺️ #GI2020 | 0.9950782 |
| craigandersn |
If you’re at #gi2020 then check out my poster: “Insights into strand-asymmetric mutational processes using segregating lesions in cancer genomes” https://t.co/rIdb9h7HPD I’ll be online to chat about #lesionsegregation and mutational processes in various asymmetric contexts. |
0.9948208 |
| acritschristoph | And we encourage questions / comments / constructive criticism / savage takedowns here or in medrxiv comments - many of us are newcomers to the viral world! Also you can see my short talk on this project on Wednesday at #GI2020, I will try to wake up for the London times | 0.9948208 |
| pathogenomenick | Evolution session up at #GI2020 - starting with @Kelley__Harris on mutation spectrum variation across the tree of life: how’s the virtual conference format going for you? I will be opening a beer at 6pm around the time of @ctitusbrown’s talk. | 0.9942157 |
| thesteinegger | PenguiN is a ProtEiN Guided Nucleotide assembler. It recovers many fold more viruses from metatranscriptomes than state of the art assemblers. Here is our #GI2020 Flash talk: https://t.co/I7Iawn9fMT Poster: https://t.co/WTEcdssJgq | 0.9938569 |
| camerongenomics | @khmiga Great #GI2020 talk! I’m really interested in your take on how we’ve gone about resolving somatic centromeric SVs (poster 17). We’re matching ~50% using hg38 repeats but surely we’d do better with a T2T reference. What approach would you recommend? | 0.9934505 |
| hdashnow | Here’s my #GI2020 lightning talk and poster. It describes the STRling algorithm and a first look at running it on 2504 genomes from the 1000g project Talk: https://t.co/I1cYFrGHdc Poster: https://t.co/l71hgLvLzt | 0.9929866 |
| SejModha | If you’ve ever wondered about #unknowns of human metagenomes, checkout my poster (P74) in today’s session at #GI2020. A short introductory talk video here: https://t.co/ejkmgJoPPk | 0.9929866 |
| ctitusbrown | Now, #GI2020 @johanneskoester talking about varlociraptor, an approach that enables a unified statistical approach to false discovery rate in variant calling. | 0.9924520 |
| pathogenomenick | Final day of #GI2020 today, starting with my favourite session: Microbes and Metagenomics, chaired by @alicecarolyn featuring @RafalMostowy, @GraceBlackwel1 and Youwen Quin ! | 0.9924520 |
8 Links
Links to GitHub, GitLab, BitBucket, Bioconductor or CRAN mentioned in Tweets.
| Name | Tweets | Retweets | Type | Link |
|---|---|---|---|---|
| bambu | 1 | 0 | GitHub | goekelab/bambu |
| chm13 | 1 | 0 | GitHub | nanopore-wgs-consortium/chm13 |
| hifiasm | 1 | 0 | GitHub | chhylp123/hifiasm |
| m6anet | 1 | 0 | GitHub | goekelab/m6anet |
| pp-sketchlib | 1 | 0 | GitHub | johnlees/pp-sketchlib |
| spacegraphcats | 1 | 0 | GitHub | spacegraphcats/spacegraphcats |
Session info
## R version 4.0.0 (2020-04-24)
## Platform: x86_64-apple-darwin17.0 (64-bit)
## Running under: macOS Catalina 10.15.6
##
## Matrix products: default
## BLAS: /Library/Frameworks/R.framework/Versions/4.0/Resources/lib/libRblas.dylib
## LAPACK: /Library/Frameworks/R.framework/Versions/4.0/Resources/lib/libRlapack.dylib
##
## locale:
## [1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] fs_1.5.0 here_0.1 kableExtra_1.2.1 knitr_1.29
## [5] magick_2.4.0 webshot_0.5.2 viridis_0.5.1 viridisLite_0.3.0
## [9] wordcloud_2.6 RColorBrewer_1.1-2 ggtext_0.1.0 ggraph_2.0.3
## [13] ggrepel_0.8.2 ggplot2_3.3.2 emo_0.0.0.9000 rvest_0.3.6
## [17] xml2_1.3.2 topicmodels_0.2-11 tidytext_0.2.5 igraph_1.2.5
## [21] stringr_1.4.0 purrr_0.3.4 forcats_0.5.0 lubridate_1.7.9
## [25] tidyr_1.1.1 dplyr_1.0.1 rtweet_0.7.0 clamour_0.1.0
##
## loaded via a namespace (and not attached):
## [1] bitops_1.0-6 usethis_1.6.1 httr_1.4.2 rprojroot_1.3-2
## [5] SnowballC_0.7.0 tools_4.0.0 backports_1.1.8 utf8_1.1.4
## [9] R6_2.4.1 colorspace_1.4-1 withr_2.2.0 tidyselect_1.1.0
## [13] gridExtra_2.3 processx_3.4.3 curl_4.3 compiler_4.0.0
## [17] cli_2.0.2 NLP_0.2-0 labeling_0.3 slam_0.1-47
## [21] scales_1.1.1 tm_0.7-7 callr_3.4.3 askpass_1.1
## [25] digest_0.6.25 rmarkdown_2.3 pkgconfig_2.0.3 htmltools_0.5.0
## [29] highr_0.8 rlang_0.4.7 rstudioapi_0.11 farver_2.0.3
## [33] generics_0.0.2 jsonlite_1.7.0 tokenizers_0.2.1 RCurl_1.98-1.2
## [37] magrittr_1.5 modeltools_0.2-23 Matrix_1.2-18 Rcpp_1.0.5
## [41] munsell_0.5.0 fansi_0.4.1 lifecycle_0.2.0 stringi_1.4.6
## [45] yaml_2.2.1 MASS_7.3-51.6 plyr_1.8.6 grid_4.0.0
## [49] parallel_4.0.0 crayon_1.3.4 lattice_0.20-41 graphlayouts_0.7.0
## [53] gridtext_0.1.1 ps_1.3.3 pillar_1.4.6 markdown_1.1
## [57] reshape2_1.4.4 stats4_4.0.0 glue_1.4.1 evaluate_0.14
## [61] vctrs_0.3.2 png_0.1-7 tweenr_1.0.1 selectr_0.4-2
## [65] gtable_0.3.0 openssl_1.4.2 polyclip_1.10-0 assertthat_0.2.1
## [69] xfun_0.16 ggforce_0.3.2 tidygraph_1.2.0 janeaustenr_0.1.5
## [73] tibble_3.0.3 ellipsis_0.3.1