biocasia2019
BioC Asia 2019
Last built: 2020-09-01 18:04:36
| Parameter | Value |
|---|---|
| hashtag | #biocasia |
| start_day | 2019-12-05 |
| end_day | 2019-12-06 |
| timezone | Australia/Sydney |
| theme | theme_light |
| accent | #3f92ac |
| accent2 | #9FC8D5 |
| kcore | 2 |
| topics_k | 6 |
| bigram_filter | 3 |
| fixed | TRUE |
| seed | 1 |
Introduction
An analysis of tweets from the #biocasia hashtag. A total of 498 tweets from 113 users were collected using the rtweet R package.
An analysis of tweets from the #biocasia hashtag for the BioC Asia conference 2019.
A total of 498 tweets from 113 users were collected using the rtweet R package.
1 Timeline
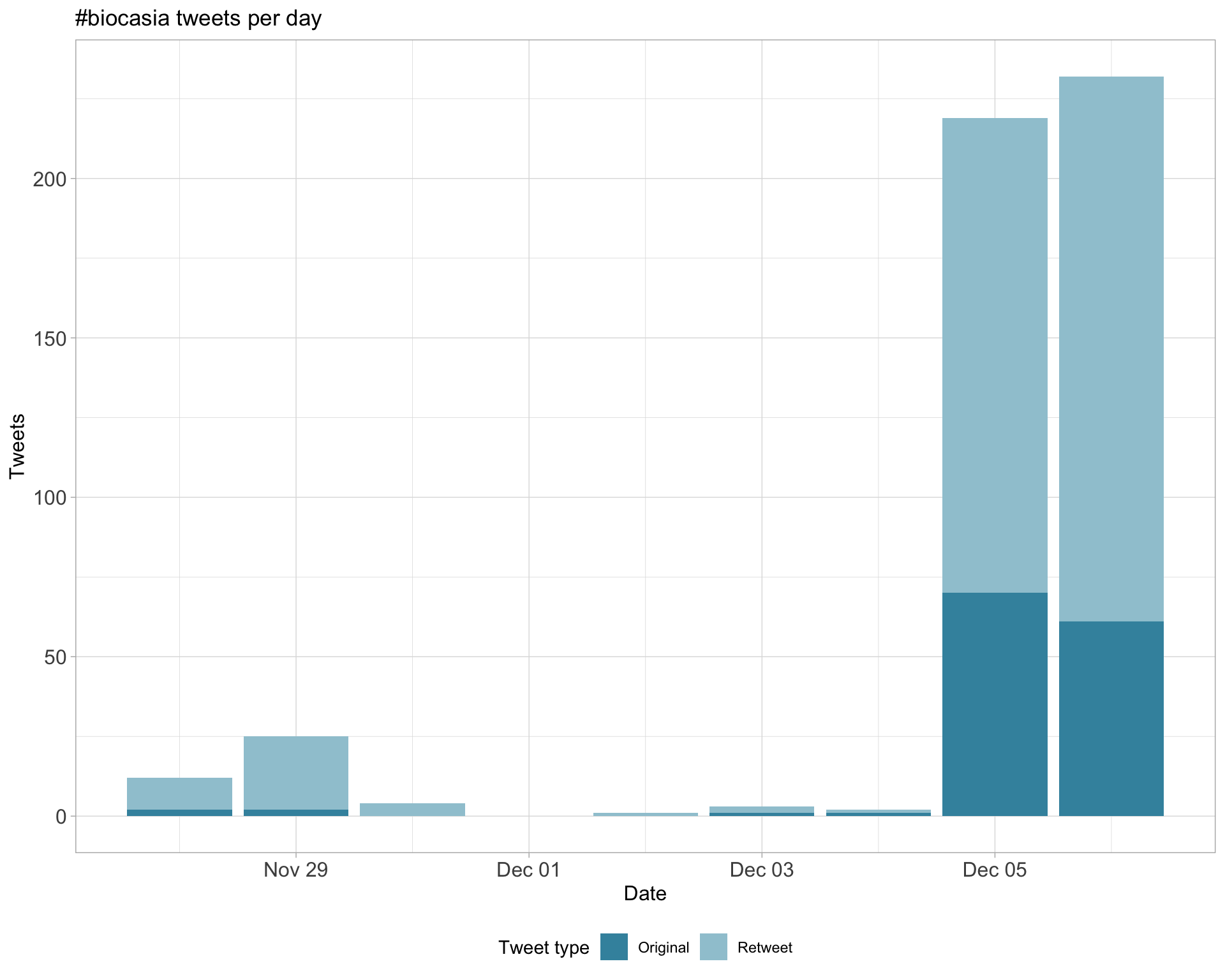
1.1 Tweets by day
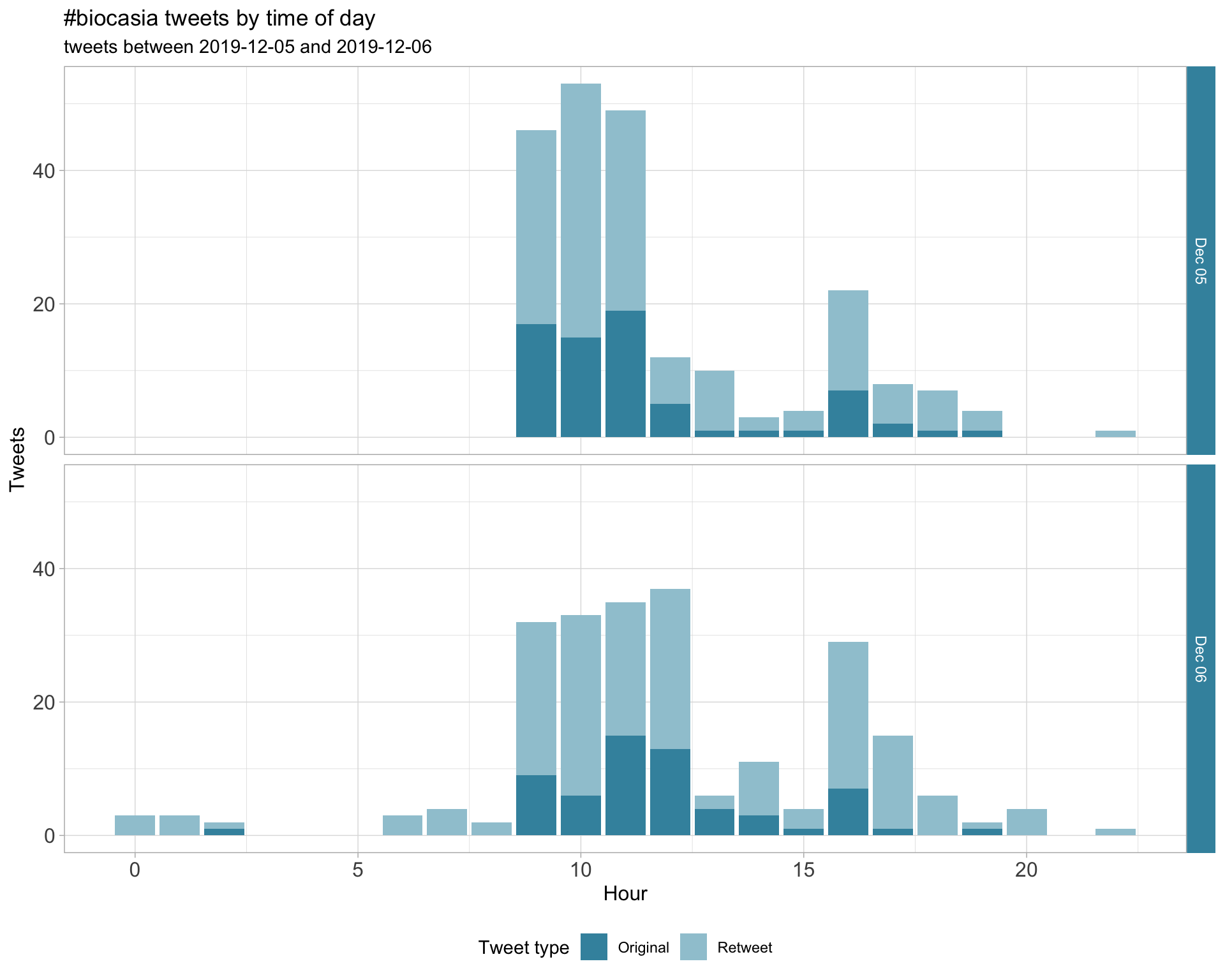
1.2 Tweets by day and time
Filtered for dates 2019-12-05 - 2019-12-06 in the Australia/Sydney timezone.
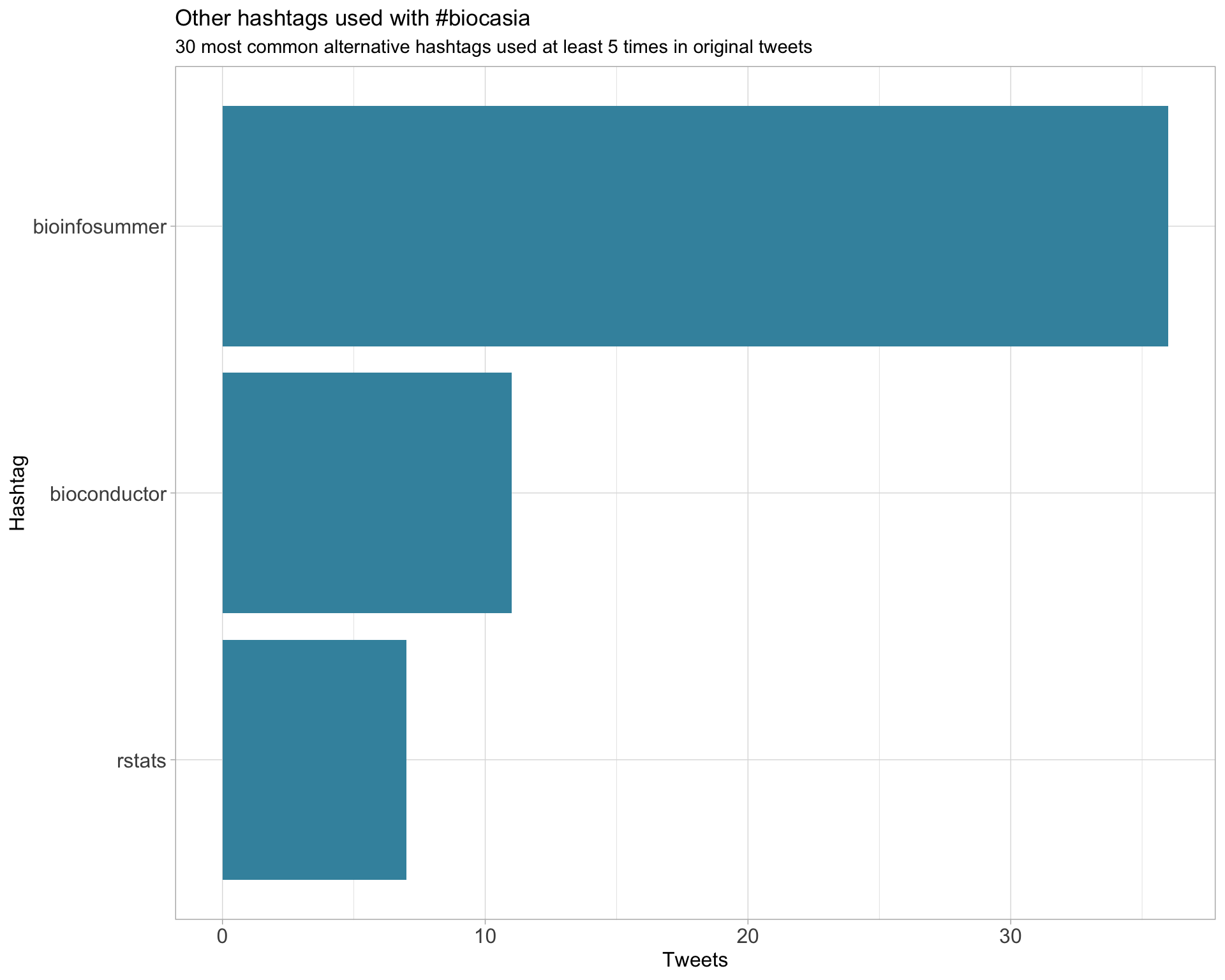
2 Users
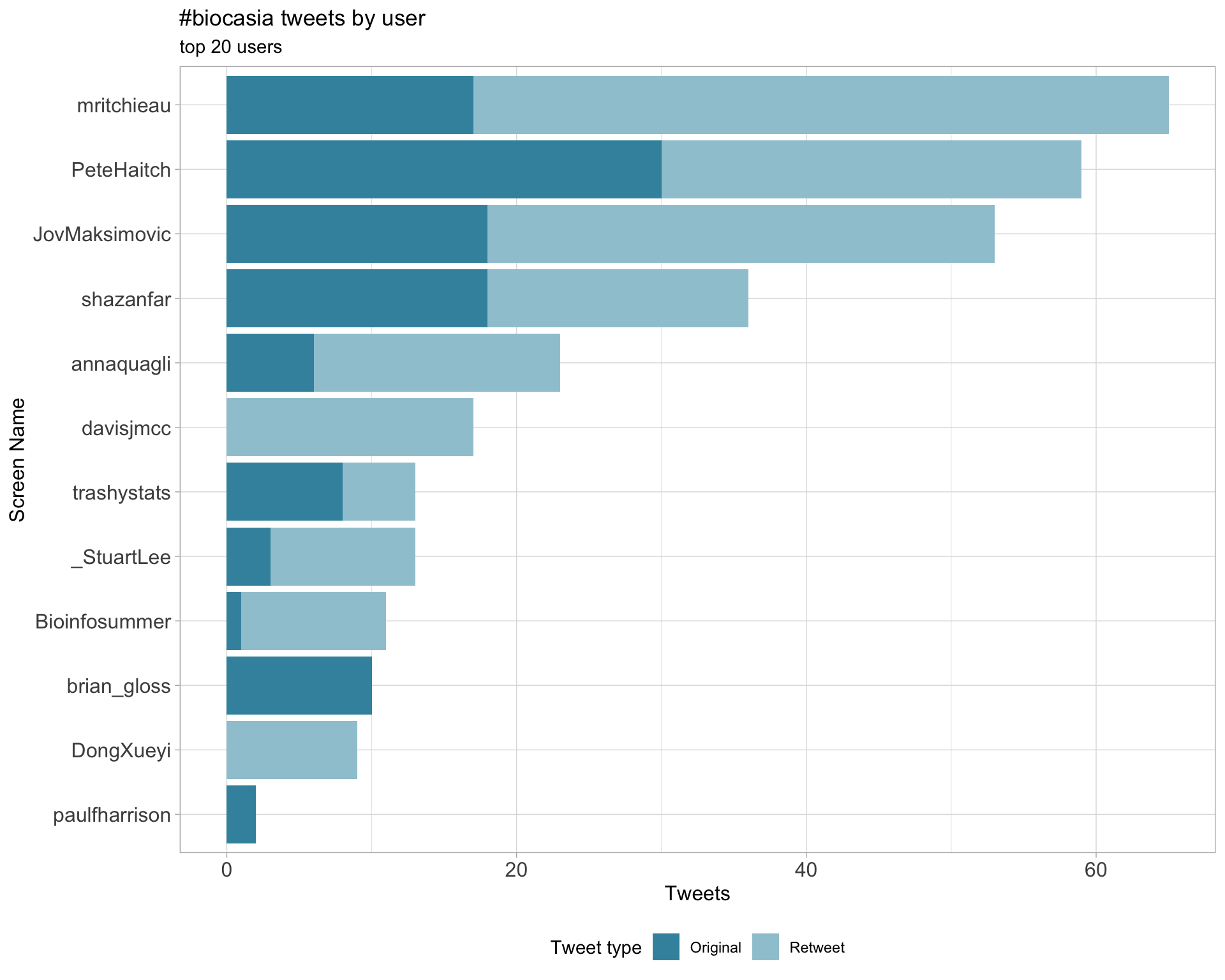
2.1 Top tweeters
Overall
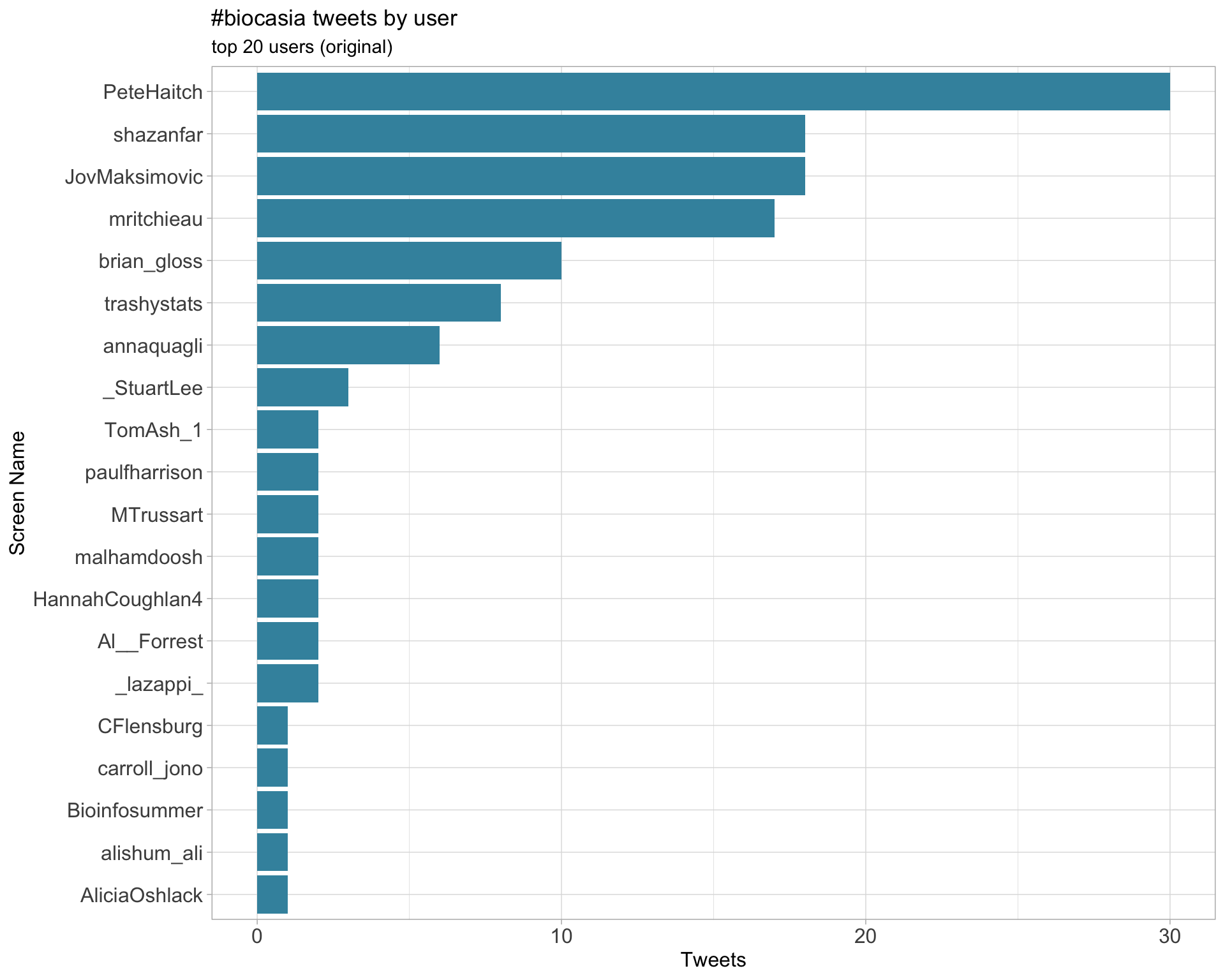
Original
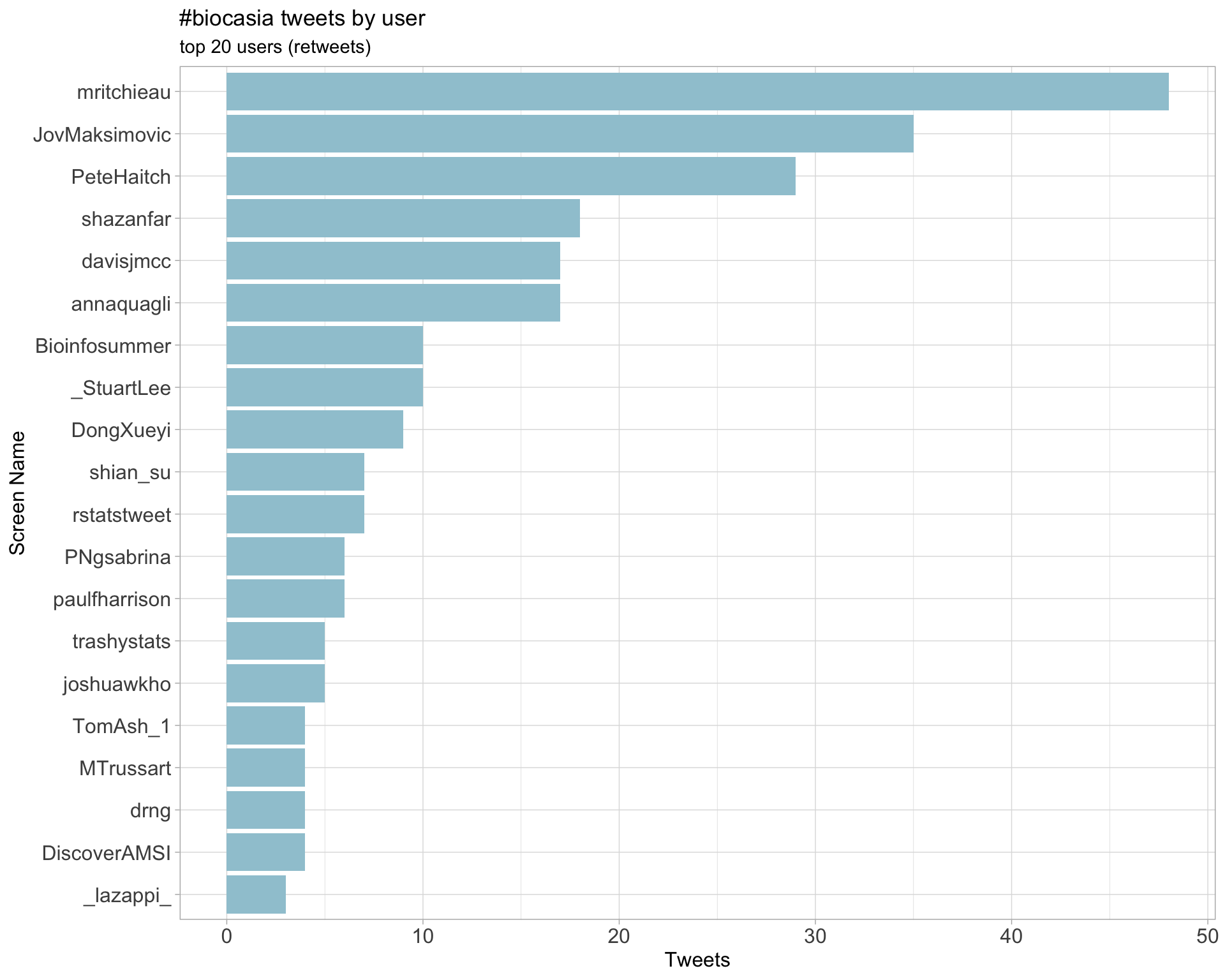
Retweets
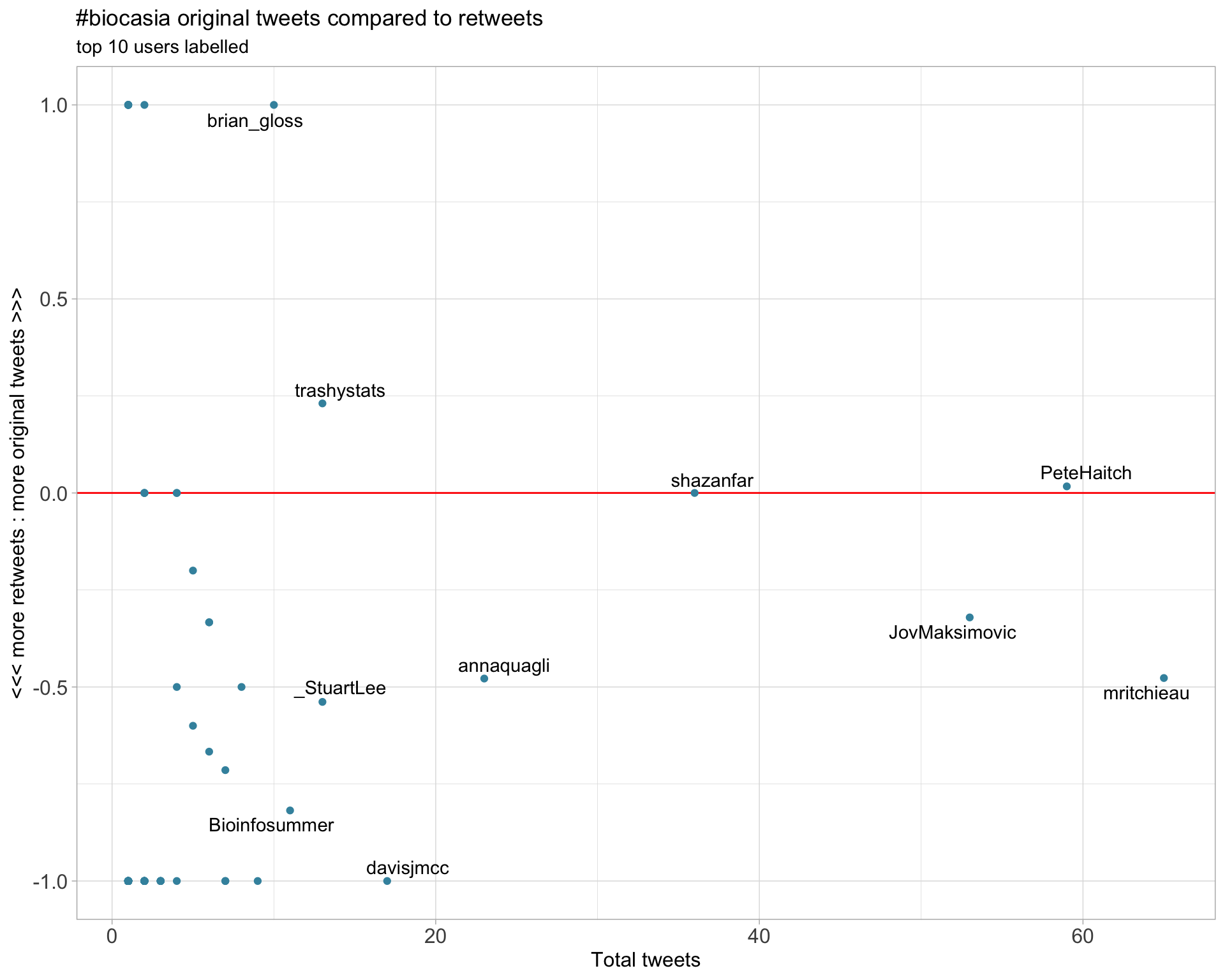
2.2 Retweet proportion
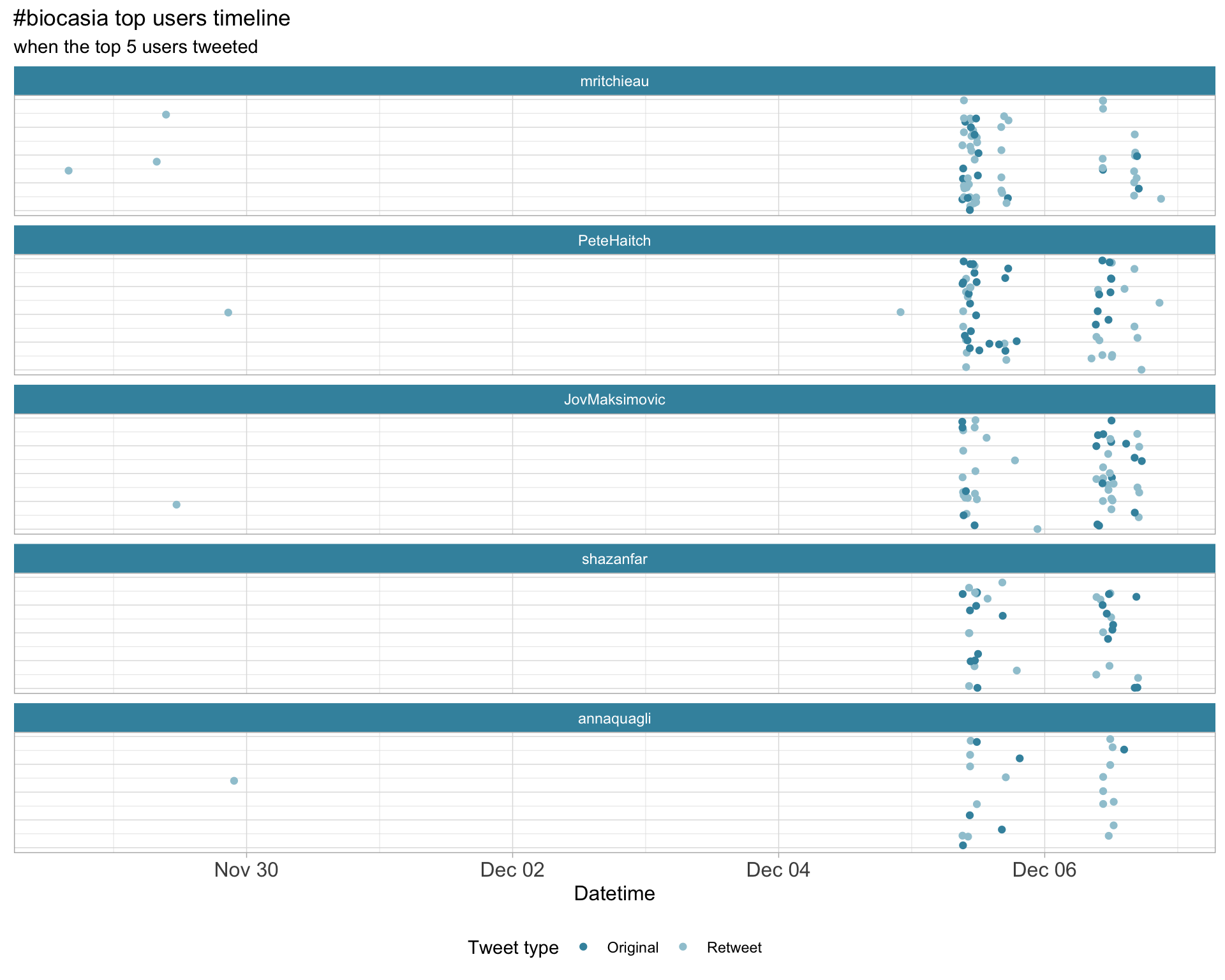
2.3 Top tweeters timeline
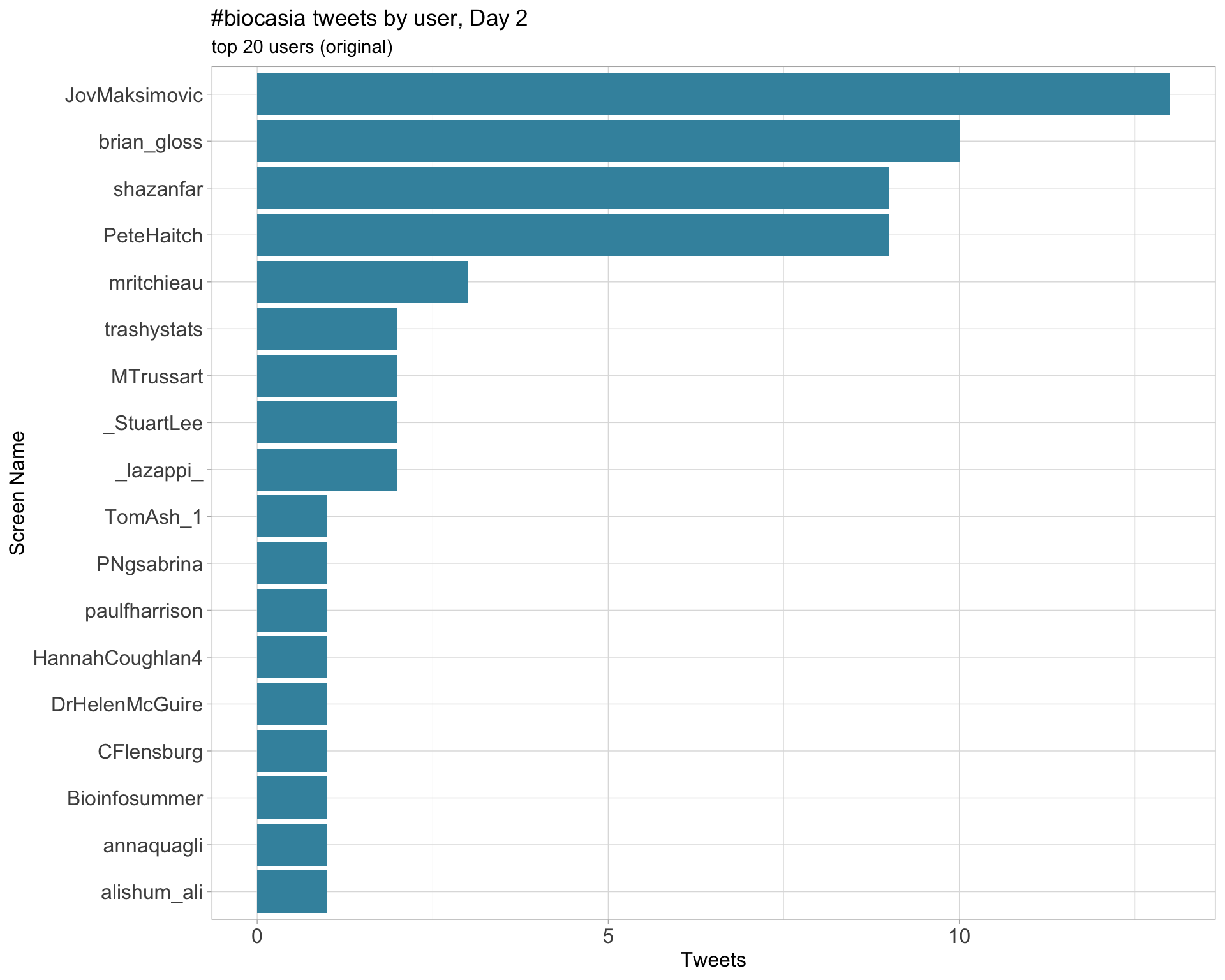
2.4 Top tweeters by day
Overall
Day 1
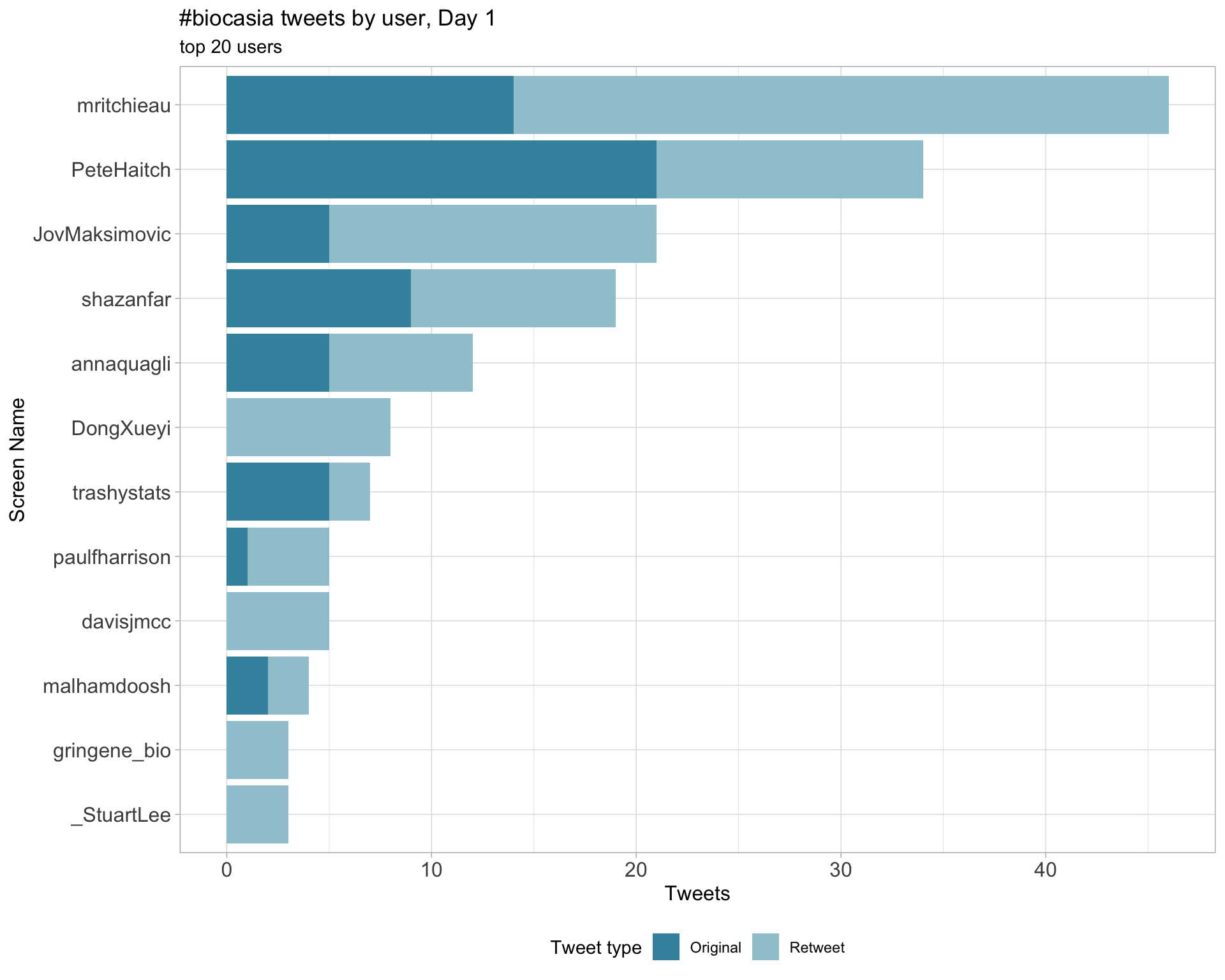
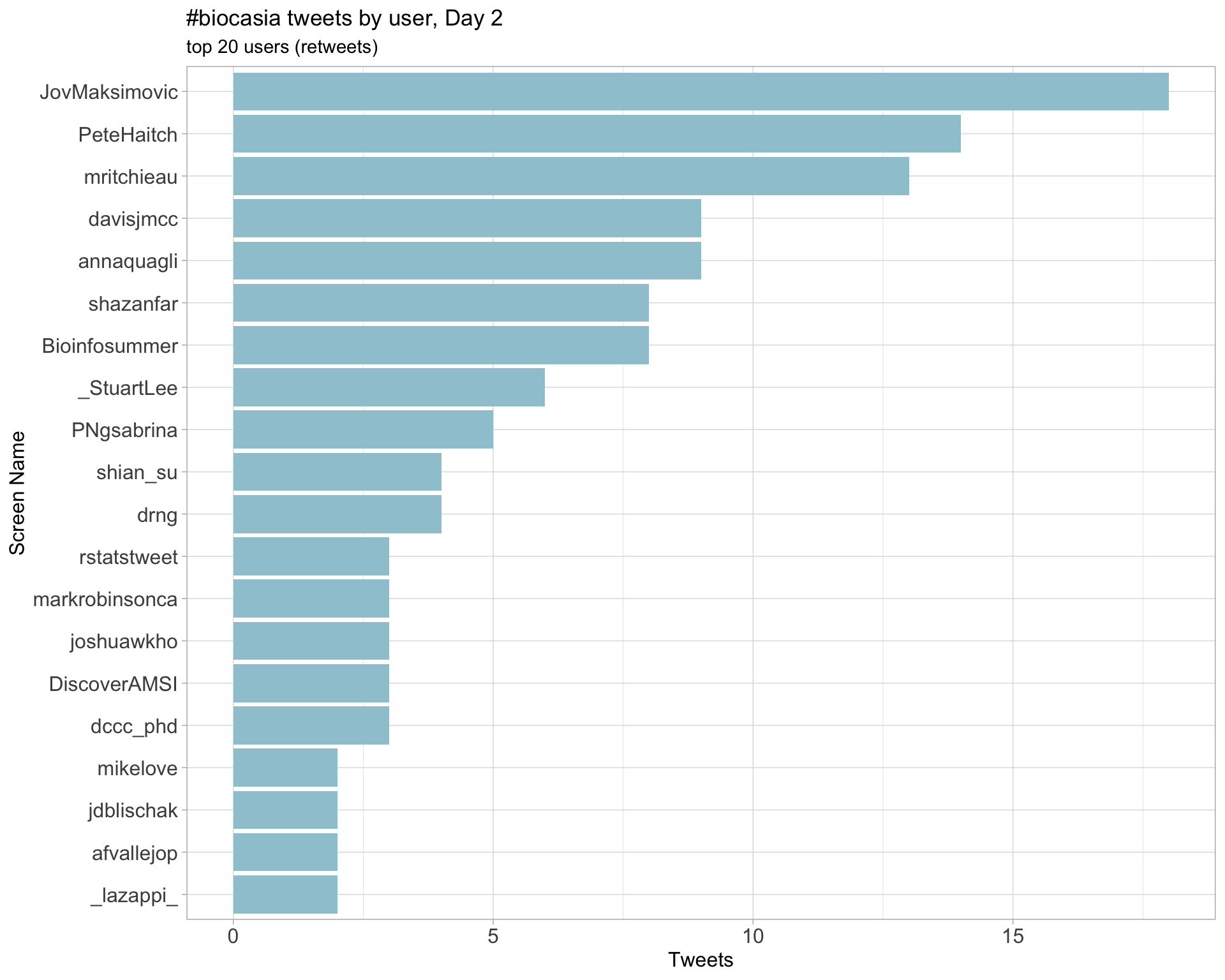
Day 2

Original
Day 1
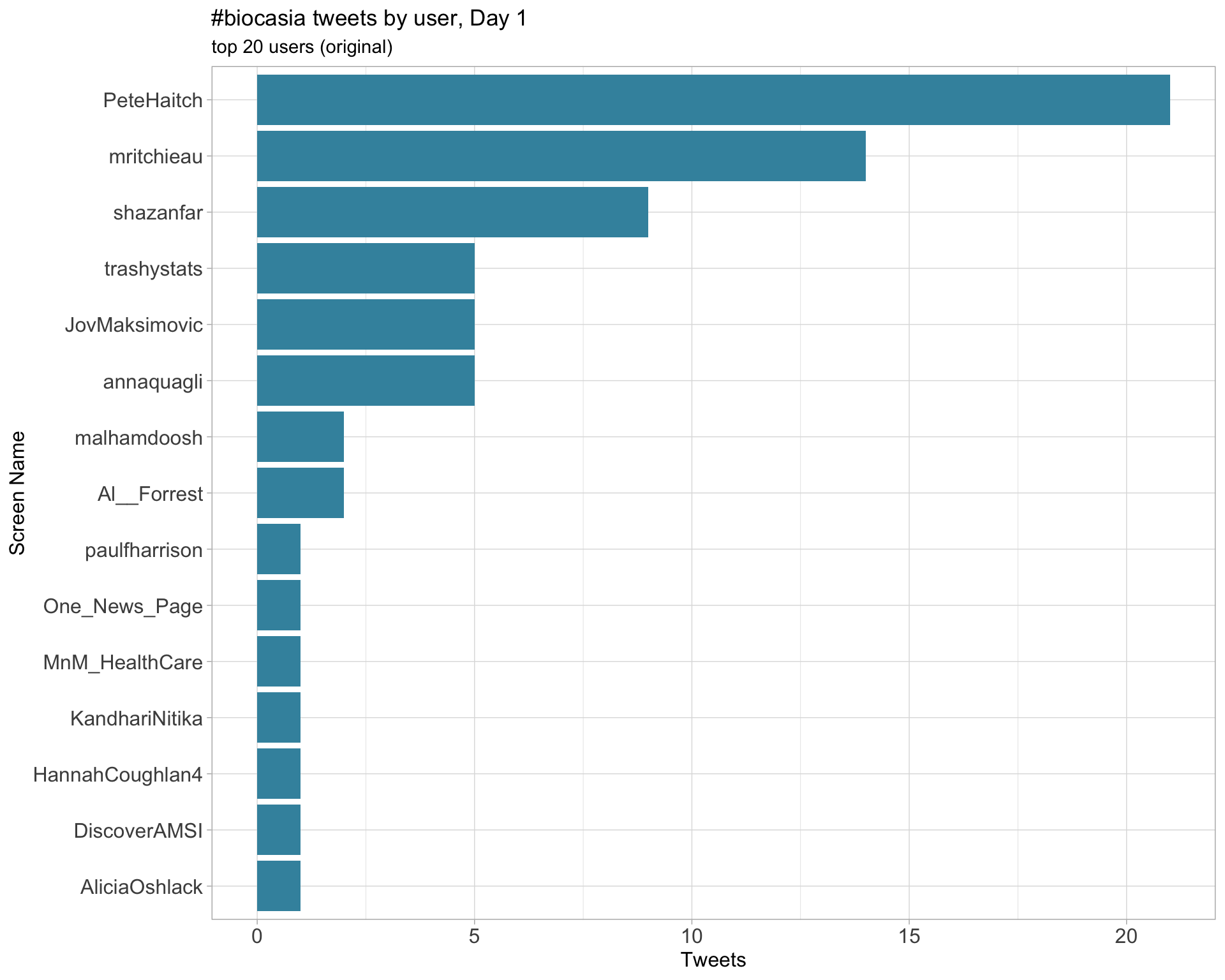
Day 2
Retweets
Day 1
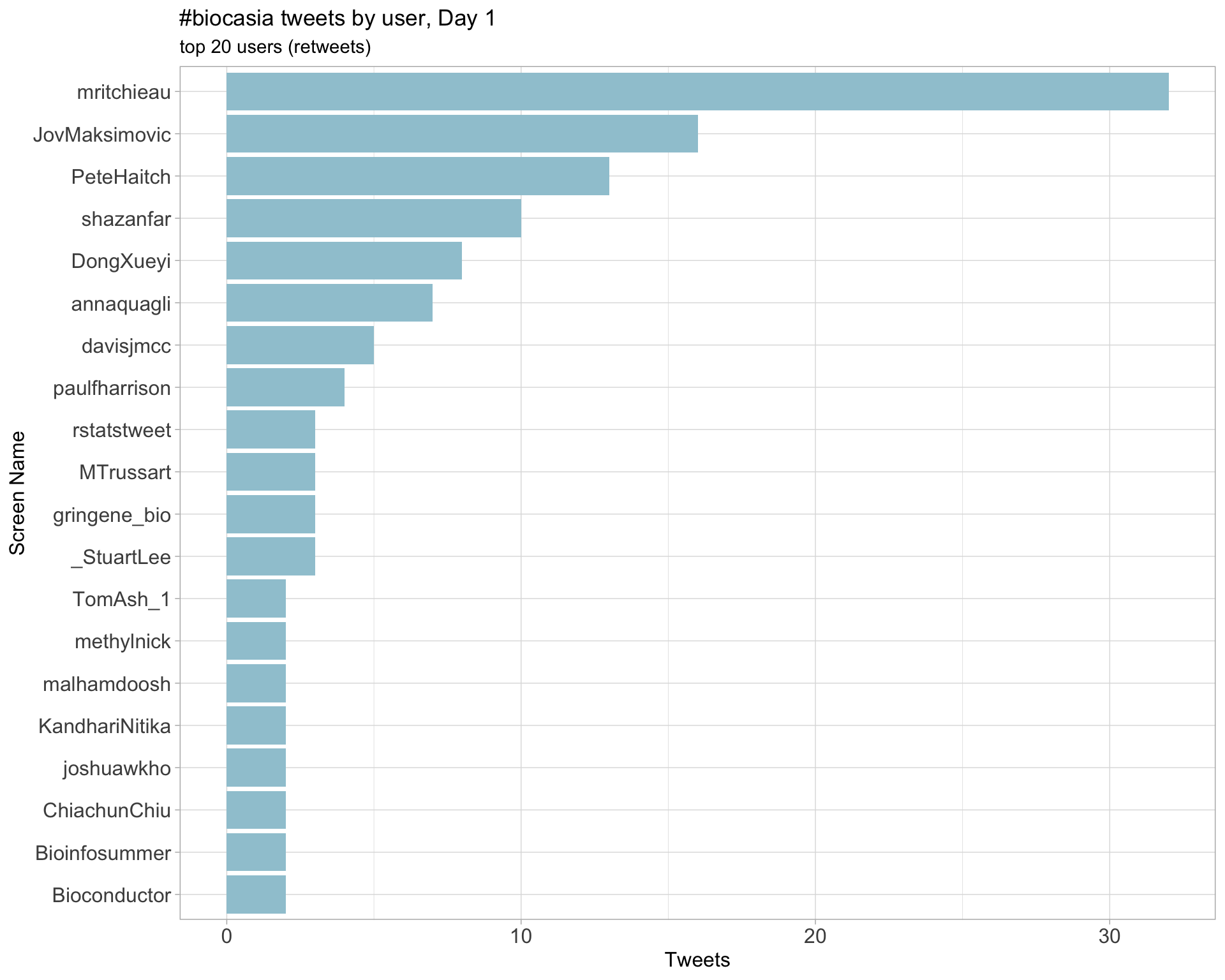
Day 2
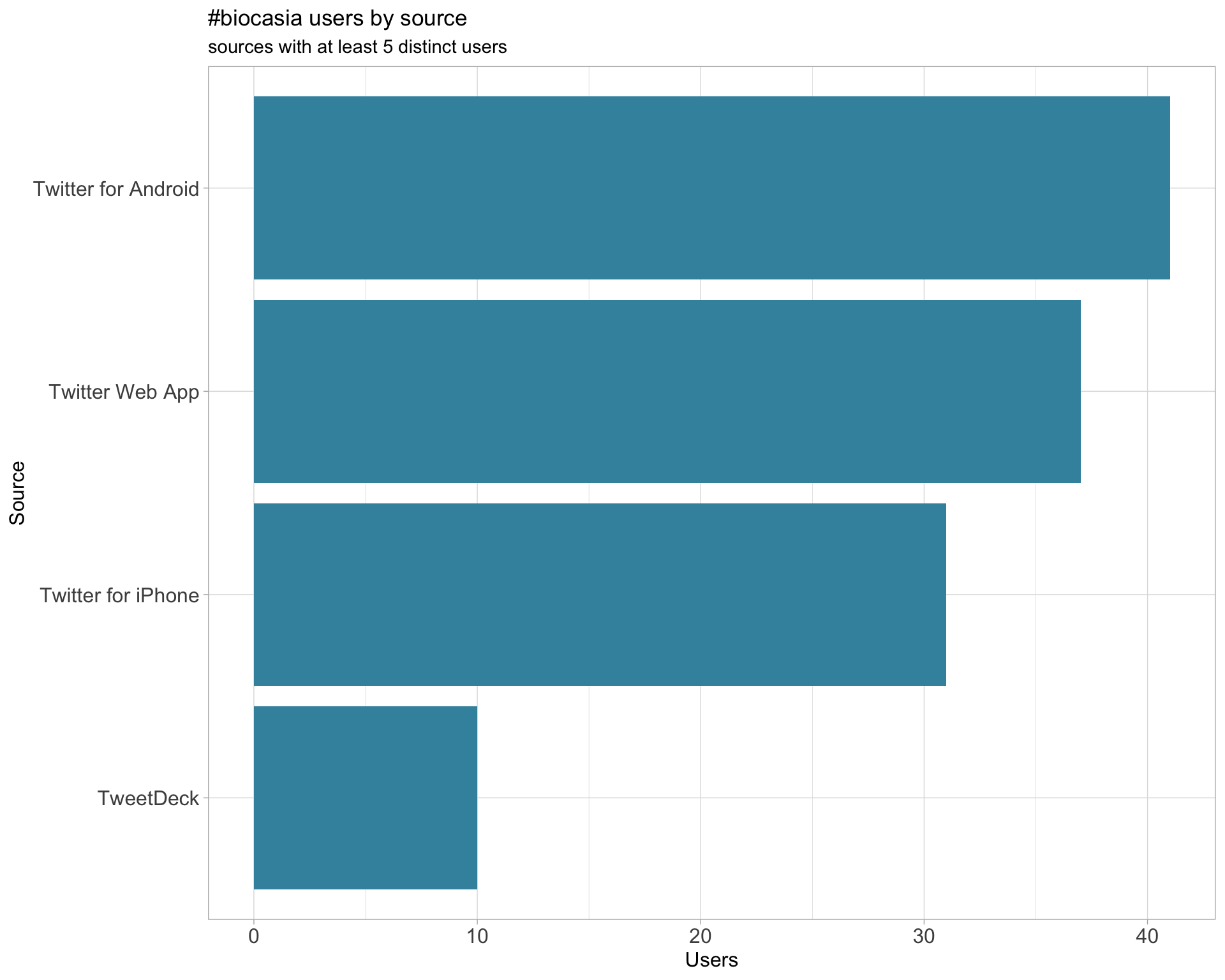
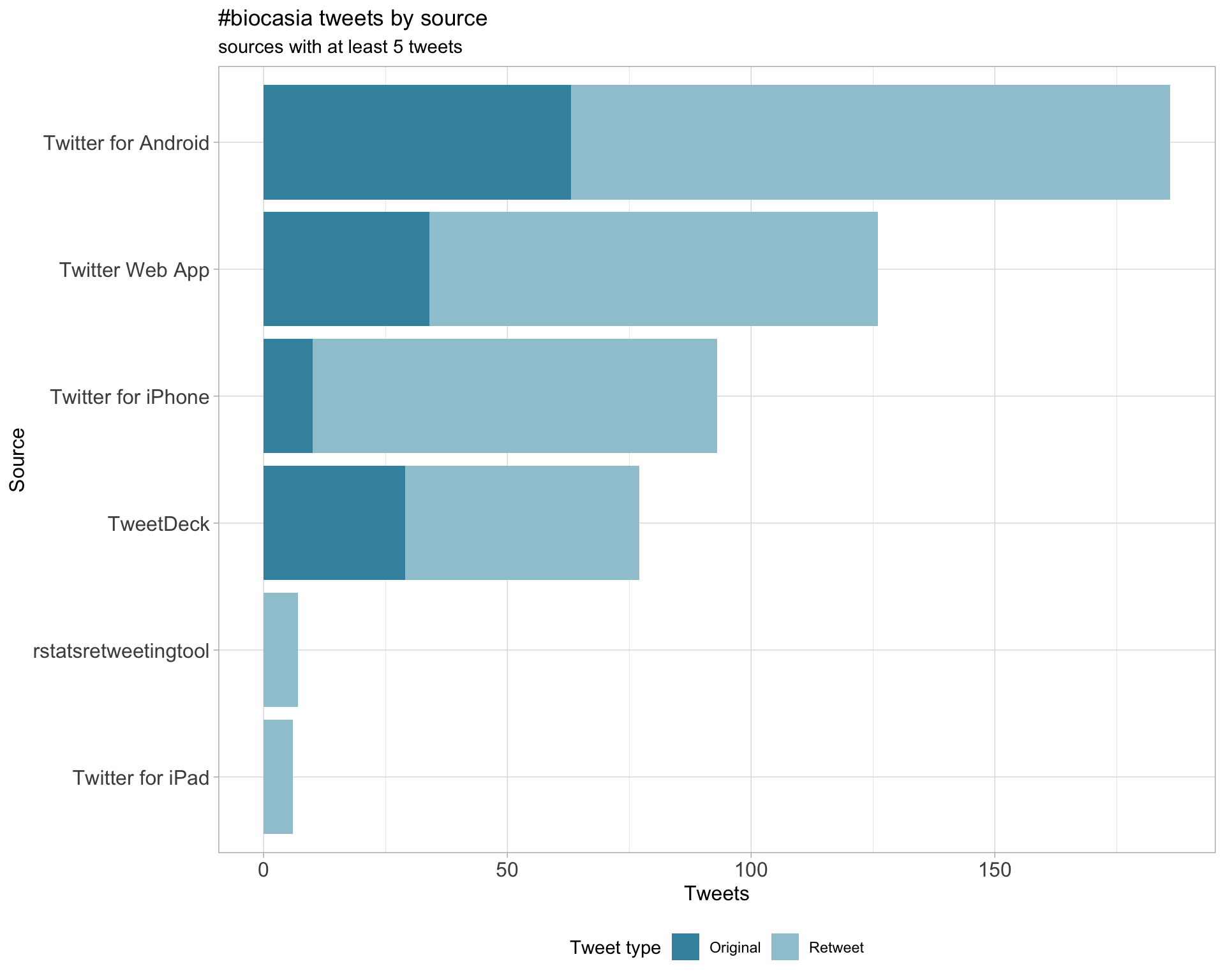
3 Sources
Users
Tweets
4 Networks
4.1 Replies
The “replies network”, composed from users who reply directly to one another, coloured by PageRank.
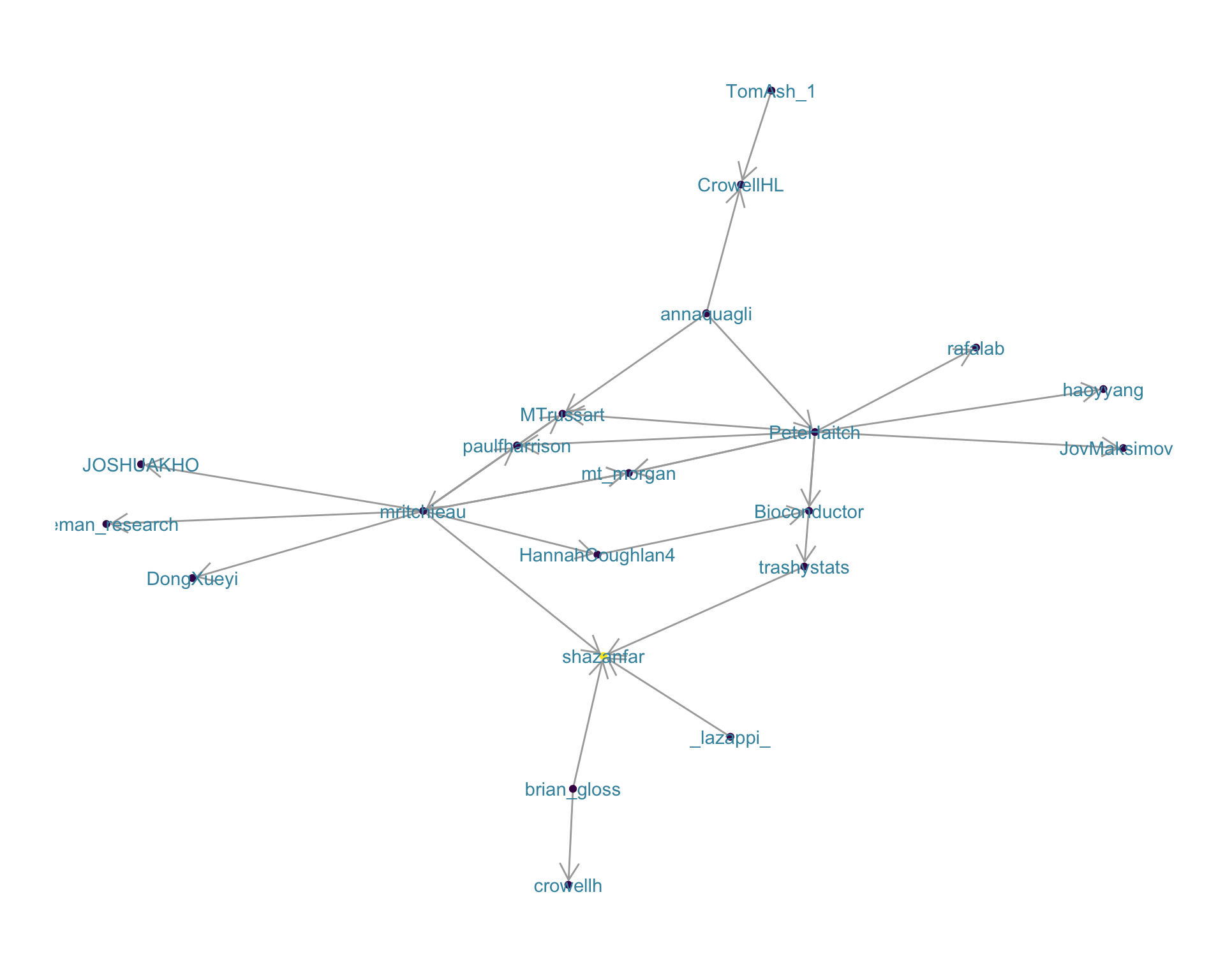
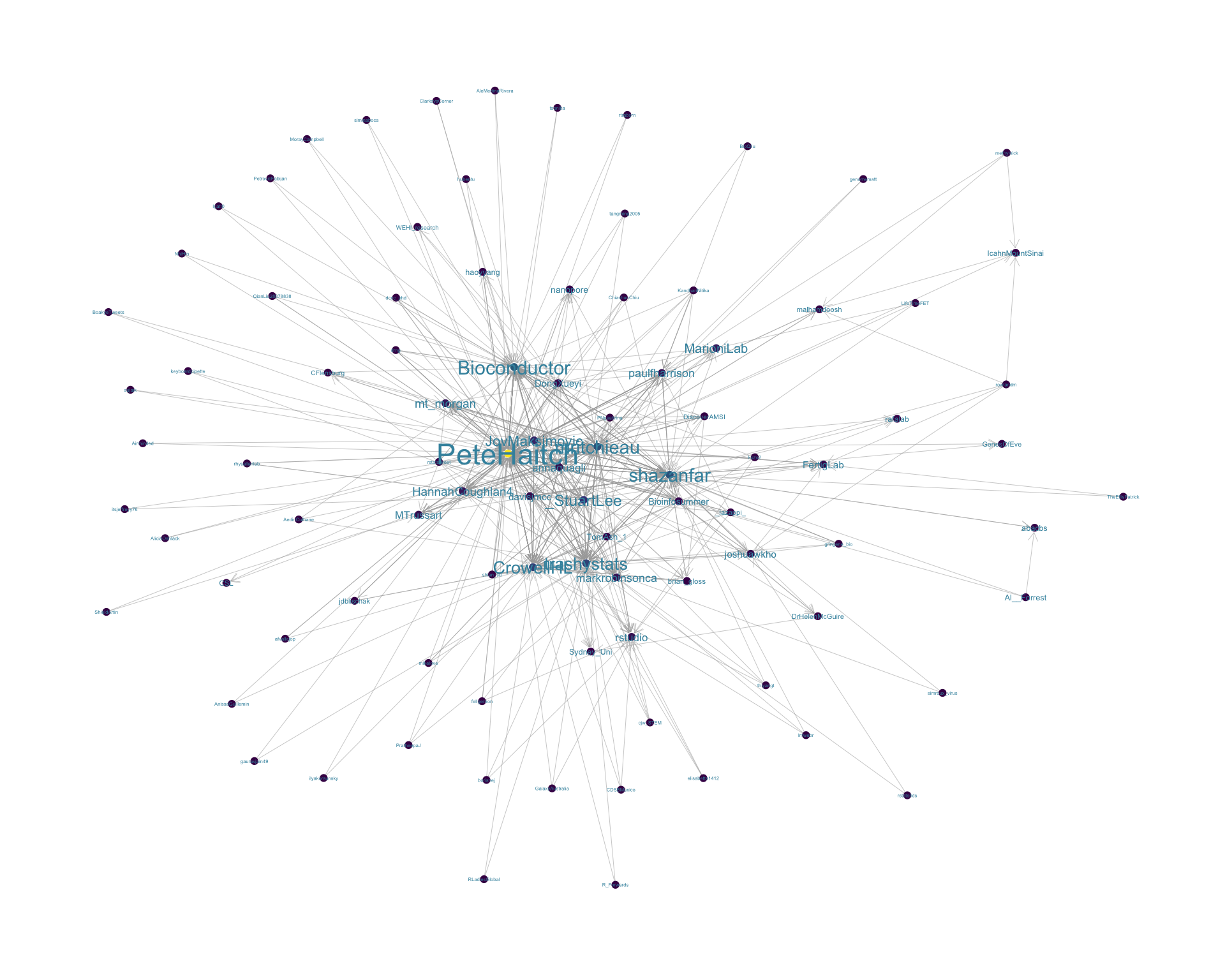
4.2 Mentions
The “mentions network”, where users mention other users in their tweets. Filtered for a k-core of 2. Node colour and size adjusted according to PageRank score.
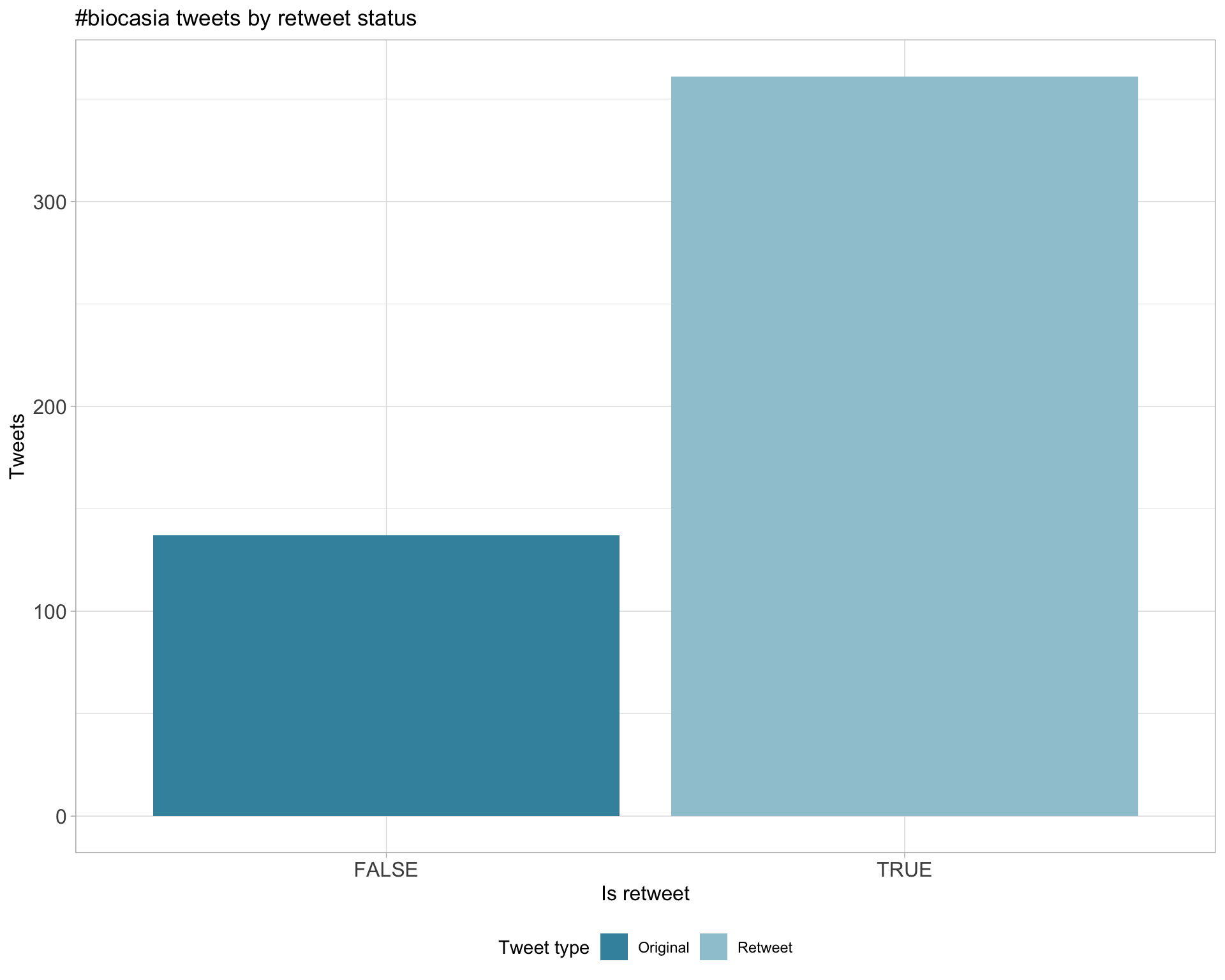
5 Tweet types
5.1 Retweets
Proportion
Count
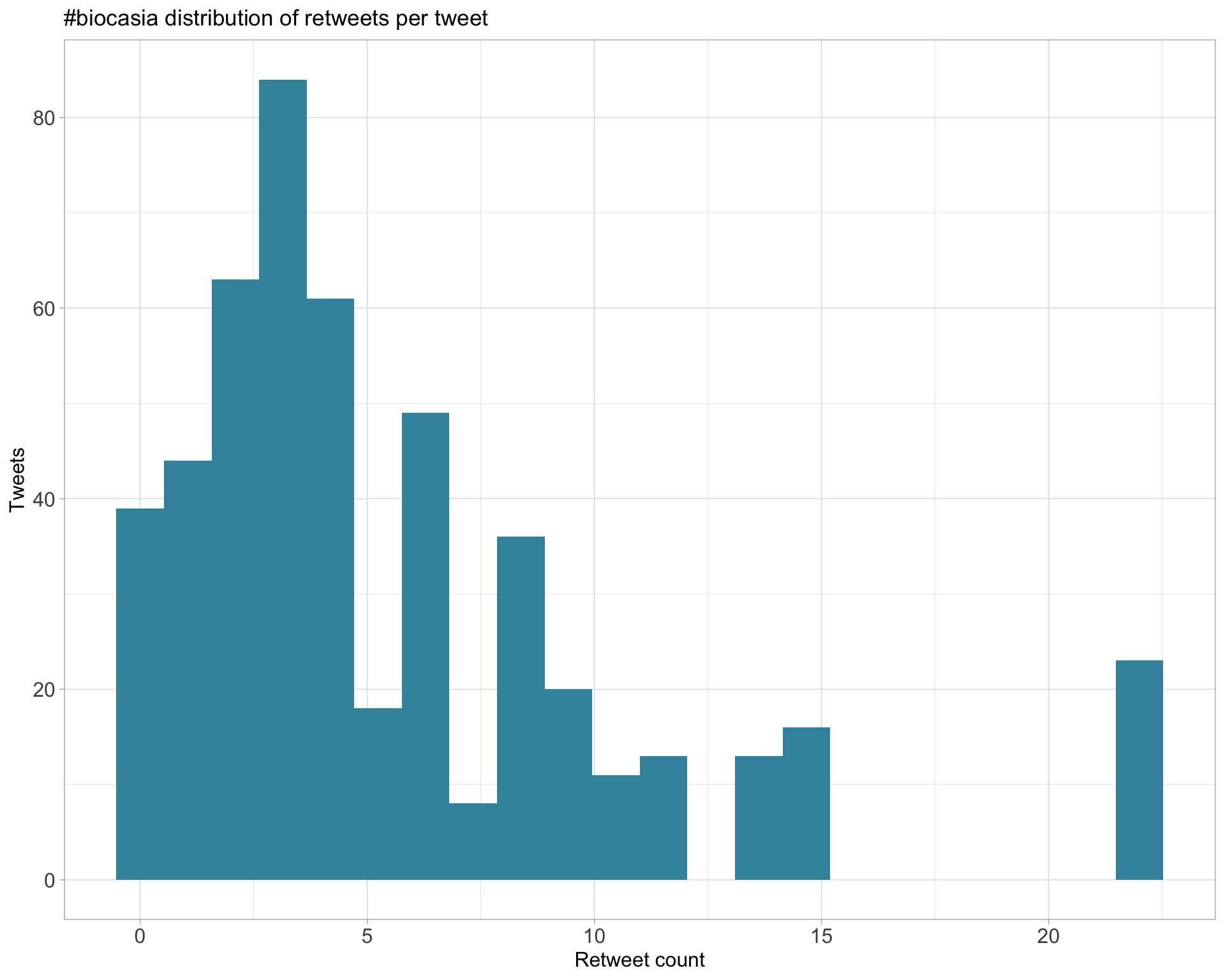
Top 10
| screen_name | text | retweet_count |
|---|---|---|
| _StuartLee | For #BiocAsia I’ve written a workflow package for exploring and combining results from RNA-seq and ATAC-seq data. First iteration is up here: https://t.co/03oRjSfThQ | 22 |
| PeteHaitch | By constructing ‘psuedobulk’ samples from scRNA-seq data, we can use tried and tested methods for differential expression analysis (limma, edgeR, DESeq2, etc) @CrowellHL benchmarked this approach along with other methods in https://t.co/SMNPfC7iyO #biocasia https://t.co/jZwcPFN13i | 15 |
| trashystats |
For #BiocAsia @PeteHaitch and me are giving a tutorial on how to make a Bioconductor package 📦 with @rstudio. You can find the tutorial here (feedback very welcome): https://t.co/y9vkKBgYSA |
14 |
| PeteHaitch | G’day from #biocasia https://t.co/f2e2XNNkmG | 12 |
| PeteHaitch | Martin Morgan making the pitch at #BioInfoSummer / #biocasia to learn @Bioconductor There’s lots of learning material available: https://t.co/xjRvliJxmb And the project is essential to lots of current research: https://t.co/qppX0pRQTO | 10 |
| PeteHaitch | There are 27 @Bioconductor workflows for common genomic analyses (https://t.co/7qUnzfWepD) Since we’re at #biocasia/#BioInfoSummer, @mritchieau is highlighting the true blue, dinky di, Aussie-developed RNAseq123 workflow (https://t.co/z5niD5vbBx) available in Chinese and English! | 9 |
| PeteHaitch | Delighted to have @CrowellHL as a keynote speaker at #biocasia! Helena and her colleagues in @markrobinsonca’s lab develop great @Bioconductor packages for CyTOF (https://t.co/ifzdnjDbLa) and scRNA-seq (https://t.co/YAYEhwEEus) data. https://t.co/dGh9rwdla7 | 9 |
| mritchieau | @shazanfar from the @MarioniLab introduces scHOT for flexible analysis of higher-order interactions in #singlecell data: shows us examples of variability, correlation and spatial analysis #biocasia. Code available at https://t.co/p8l2m21jqt https://t.co/41Nbf73QtU | 8 |
| PeteHaitch | @JovMaksimovic develops the missMethyl package which has the best logo in the business #biocasia https://t.co/JFMQdiJUoa https://t.co/D5uXSQNtJB | 8 |
| PeteHaitch | PuXue Qiao sharing the resources she, along with Ruqian Lyu and @davisjmcc, developed for a 2-day scRNA-seq course https://t.co/sq2q2HgEA6 #biocasia | 8 |
Most retweeted

5.2 Likes
Proportion
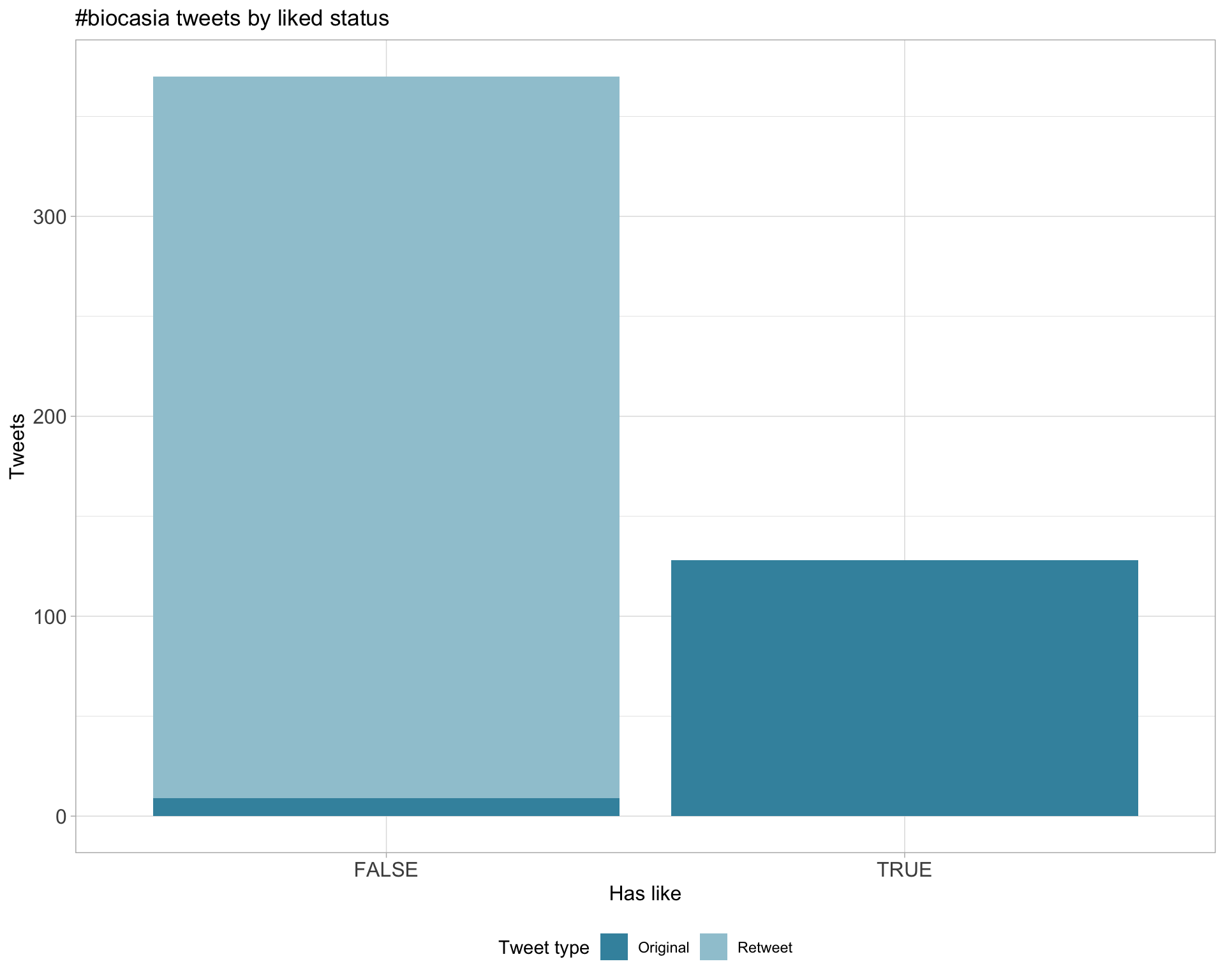
Count
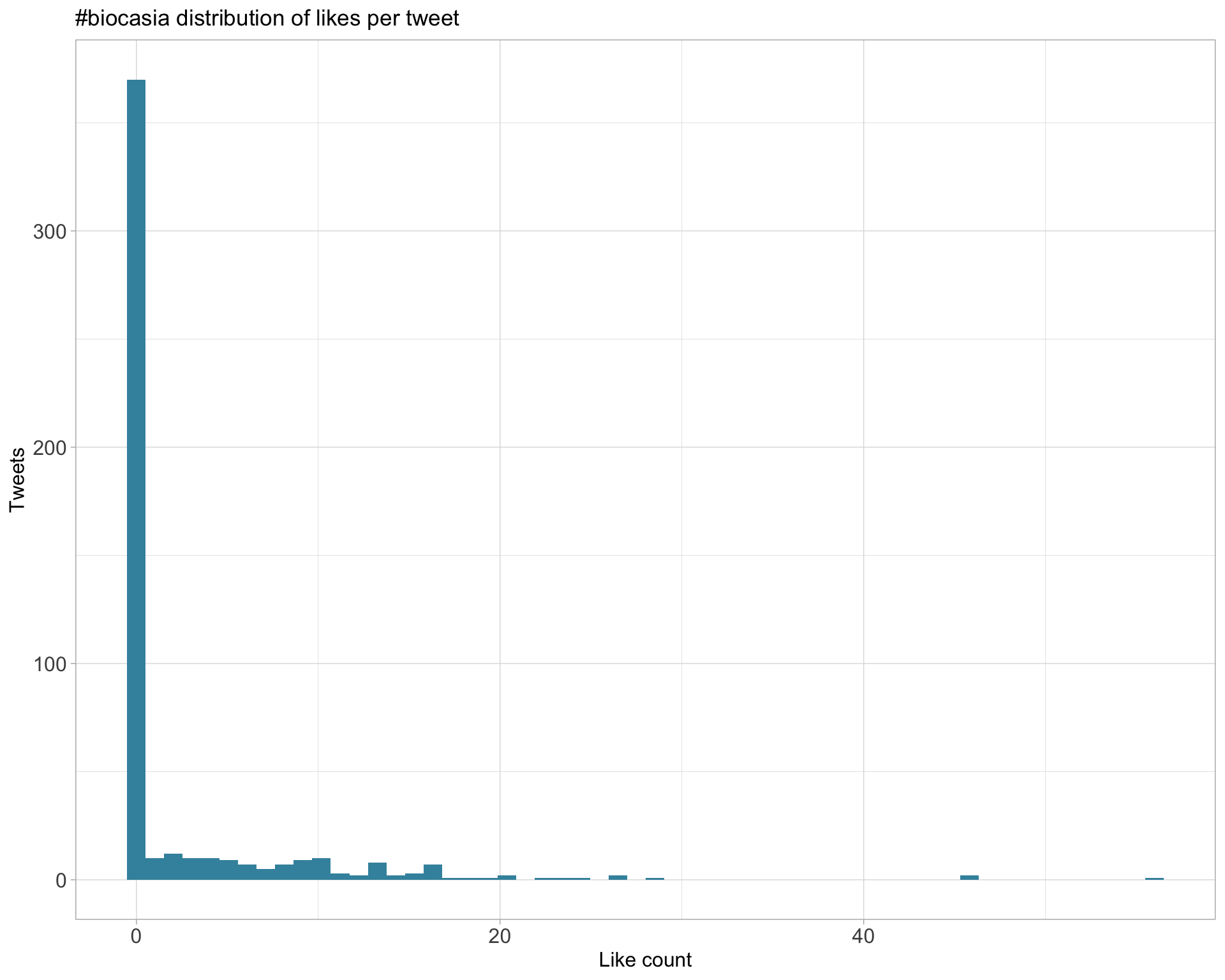
Top 10
| screen_name | text | favorite_count |
|---|---|---|
| _StuartLee | For #BiocAsia I’ve written a workflow package for exploring and combining results from RNA-seq and ATAC-seq data. First iteration is up here: https://t.co/03oRjSfThQ | 56 |
| PeteHaitch | G’day from #biocasia https://t.co/f2e2XNNkmG | 46 |
| PeteHaitch | By constructing ‘psuedobulk’ samples from scRNA-seq data, we can use tried and tested methods for differential expression analysis (limma, edgeR, DESeq2, etc) @CrowellHL benchmarked this approach along with other methods in https://t.co/SMNPfC7iyO #biocasia https://t.co/jZwcPFN13i | 46 |
| trashystats | What a great keynote!!! It is quite unbelievable that @CrowellHL is only in her first year of her PhD. #biocasia Outstanding. https://t.co/DyR64Qx5ou | 29 |
| PeteHaitch | Charity Law, developer of voom, introducing an extension to voom to handle group-specific variance estimation and how she’s thinking about adapting it to scRNA-seq differential expression analysis #biocasia https://t.co/HOguf3wWpl | 26 |
| trashystats |
For #BiocAsia @PeteHaitch and me are giving a tutorial on how to make a Bioconductor package 📦 with @rstudio. You can find the tutorial here (feedback very welcome): https://t.co/y9vkKBgYSA |
26 |
| CrowellHL | Off to 🇦🇺 for some Bioinformatics fun 👩🏻💻, to talk & teach about CyTOF at #BiocAsia / BioInfoSummer in Sydney, and explore other parts of AUS. Stressed, grateful and excited! - Now looking forward to 20h of reading & movies ✈️😴 That completes my #BiocBingo 🥳 | 24 |
| PeteHaitch | It takes 5+ bioinformaticians to get the bloody projector working, thanks to @JovMaksimovic for being willing to improvise! Excited to hear her work on gene set testing for differentially methylated regions #biocasia https://t.co/R8OUlu4Hy6 | 23 |
| PeteHaitch | You know it’s a @DongXueyi talk when Pokemon make an appearance. Xueyi is presenting her work on RNA-seq analysis of @nanopore data #biocasia https://t.co/PblubdtTSH | 22 |
| PeteHaitch | Lots of people in the house to learn about @Bioconductor from the leader of the project, Martin Morgan at #biocasia #BioInfoSummer https://t.co/lwPLmrBJJM | 20 |
Most likes
5.3 Quotes
Proportion
Count
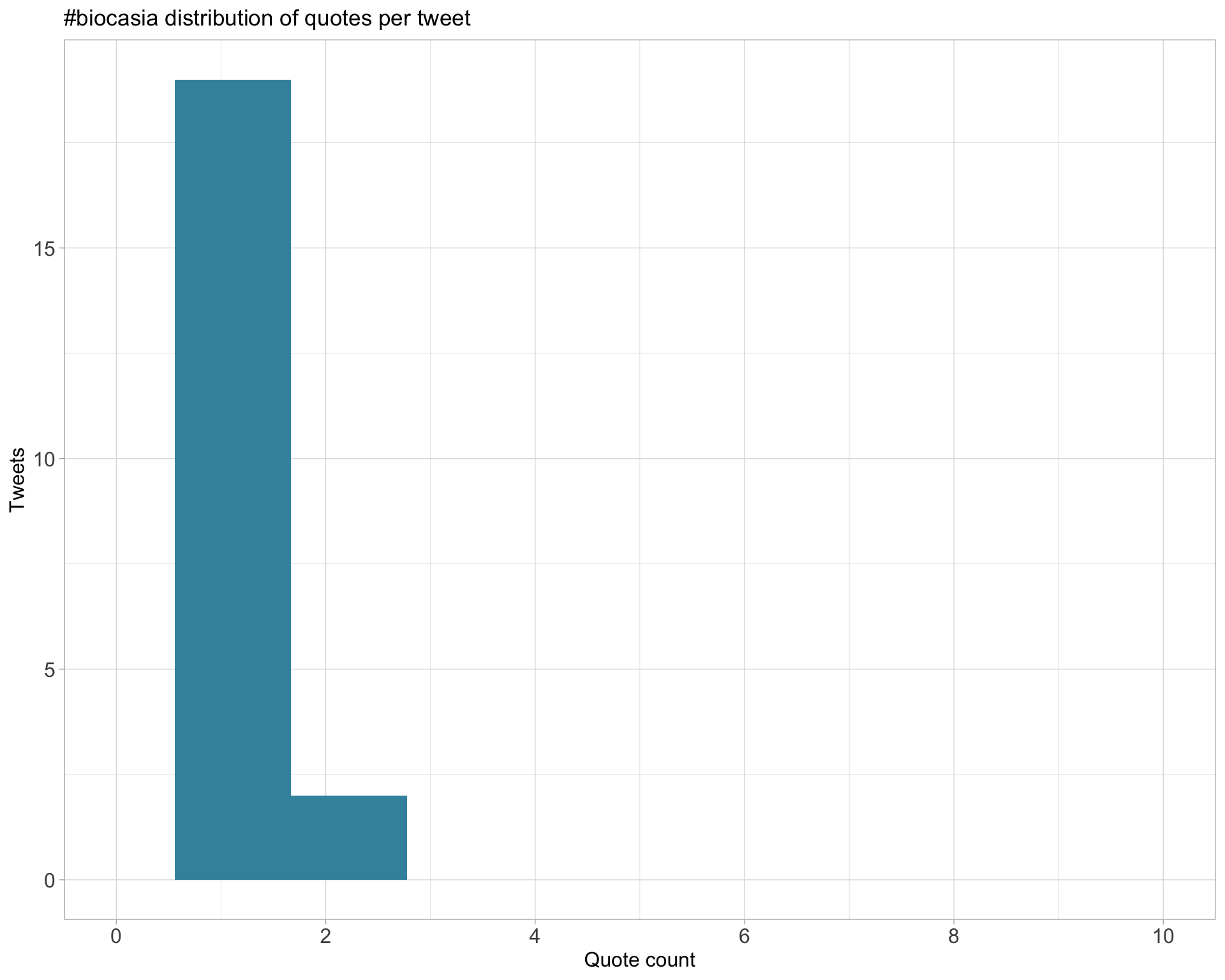
Top 10
| screen_name | text | quote_count |
|---|---|---|
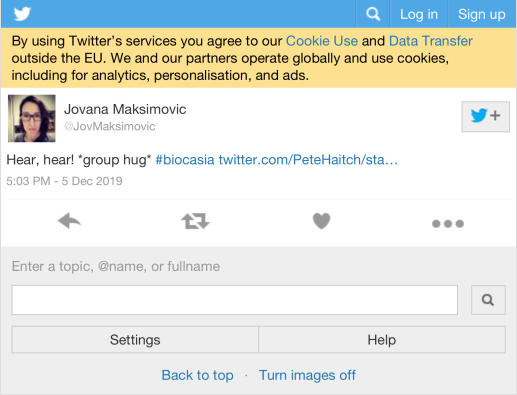
| JovMaksimovic | Hear, hear! group hug #biocasia https://t.co/OCvPxeCRj8 | 2 |
| MTrussart | Wonderful talk from @CrowellHL at #BiocAsia showcasing #Catalyst and #diffcyt #R packages, i loved her enthusiasm and emotion towards the #Bioconductor community! https://t.co/DmF10cf1gC | 2 |
| mritchieau | So many cool papers @FertigLab @GenesOfEve and colleagues to learn more about these methods e.g. https://t.co/OsJH622gAo and https://t.co/woblN8CxhC #biocasia https://t.co/7NhkR0zyuG | 2 |
| JovMaksimovic | Final talk for #BiocAsia #BioInfoSummer https://t.co/YGwlhBuLlk | 2 |
| joshuawkho | Please come join me at the #BioCAsia conference! https://t.co/RVzv1jvCzz | 1 |
| mritchieau | #biocasia https://t.co/q4IjB0zb27 | 1 |
| JovMaksimovic | I really need to get onto the @Bioconductor slack #biocasia https://t.co/GffPVaX5F8 | 1 |
| HannahCoughlan4 | Thanks for the opportunity to present my work! #biocasia https://t.co/g9i3Abkm3E | 1 |
| trashystats | This was a most impressive undergrad talk. Beautiful visualizations! #biocasia https://t.co/DZeUiqexlu | 1 |
| mritchieau | #biocasia https://t.co/DhQnm7H9md | 1 |
Most quoted
6 Media
Proportion
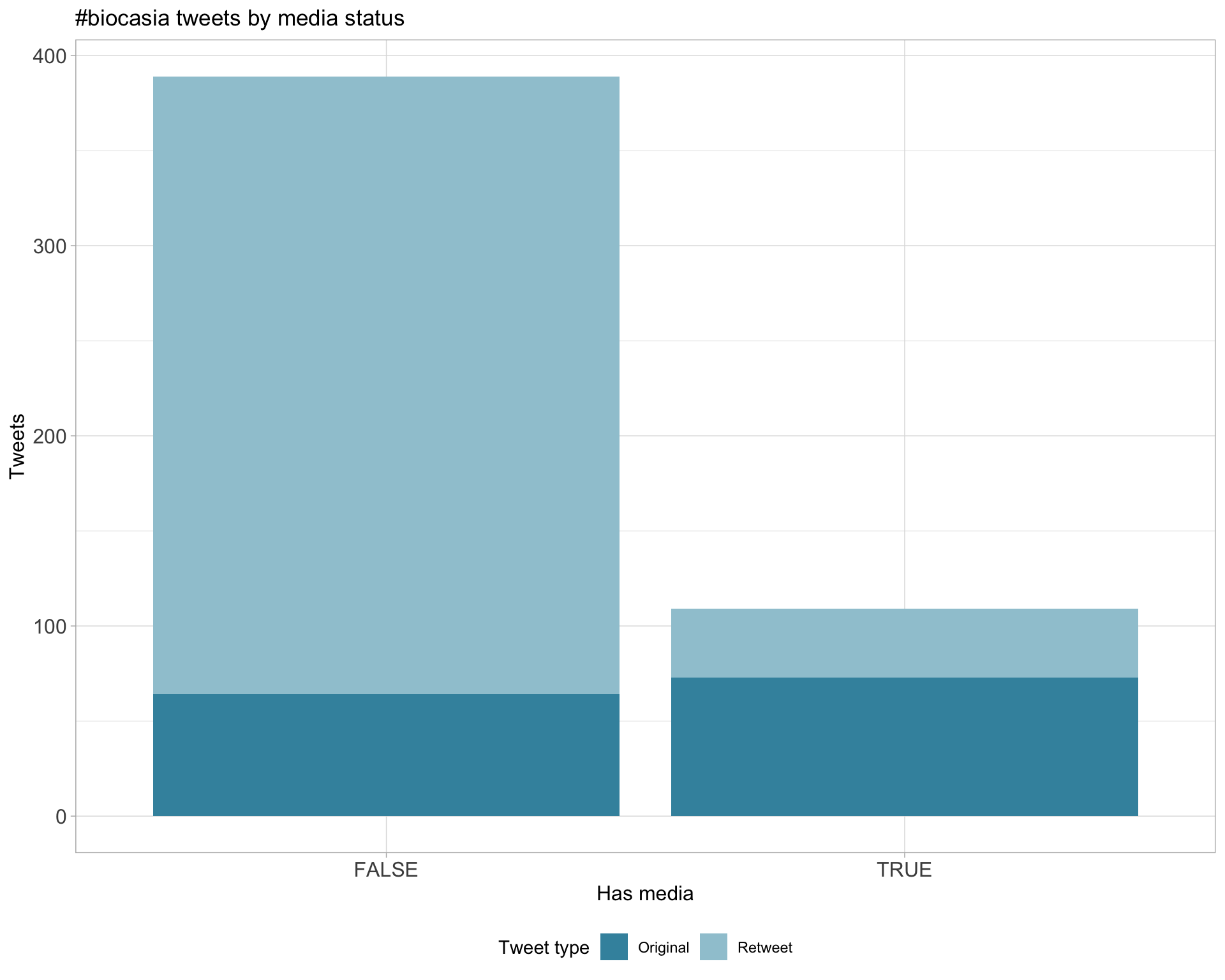
Top 10
| screen_name | text | favorite_count |
|---|---|---|
| PeteHaitch | G’day from #biocasia https://t.co/f2e2XNNkmG | 46 |
| PeteHaitch | By constructing ‘psuedobulk’ samples from scRNA-seq data, we can use tried and tested methods for differential expression analysis (limma, edgeR, DESeq2, etc) @CrowellHL benchmarked this approach along with other methods in https://t.co/SMNPfC7iyO #biocasia https://t.co/jZwcPFN13i | 46 |
| PeteHaitch | Charity Law, developer of voom, introducing an extension to voom to handle group-specific variance estimation and how she’s thinking about adapting it to scRNA-seq differential expression analysis #biocasia https://t.co/HOguf3wWpl | 26 |
| PeteHaitch | It takes 5+ bioinformaticians to get the bloody projector working, thanks to @JovMaksimovic for being willing to improvise! Excited to hear her work on gene set testing for differentially methylated regions #biocasia https://t.co/R8OUlu4Hy6 | 23 |
| PeteHaitch | You know it’s a @DongXueyi talk when Pokemon make an appearance. Xueyi is presenting her work on RNA-seq analysis of @nanopore data #biocasia https://t.co/PblubdtTSH | 22 |
| PeteHaitch | Lots of people in the house to learn about @Bioconductor from the leader of the project, Martin Morgan at #biocasia #BioInfoSummer https://t.co/lwPLmrBJJM | 20 |
| PeteHaitch | Exciting talk by @shazanfar bringing scHOT (https://t.co/1NSzfqWwLe) to spatial transcriptomics, the cool new kid on the single cell block #biocasia https://t.co/4SsGLInsmk | 20 |
| annaquagli | Thanks to @trashystats for the detailed documentation and workshop on making @Bioconductor 📦 . Did you know that you can go inside the R/myfun.R -> click on ‘Code’ -> ‘Insert Roxygen Skeleton’ ?🤯 #BiocAsia https://t.co/GDDjQW9Gny https://t.co/urkJfLMP7v | 19 |
| PeteHaitch | The “boring to some people but I think is really cool result” (@CrowellHL’s words, but I concur) is that running edgeR or limma on the summed counts works best and is many, many times faster and simpler than running a full-blown mixed model in their experience #biocasia https://t.co/YdYzI7UnpJ | 18 |
| shazanfar | #biocasia #bioinfosummer @trashystats addressing the issue of overplotting of many many coloured points in a scatterplot, with Bioconductor package schex (pron “ess-see-hex” 🤣). Saskia is super cool under the live demo pressure, well done!! https://t.co/mywX9y9MMN | 17 |
6.1 Most liked image

7 Tweet text
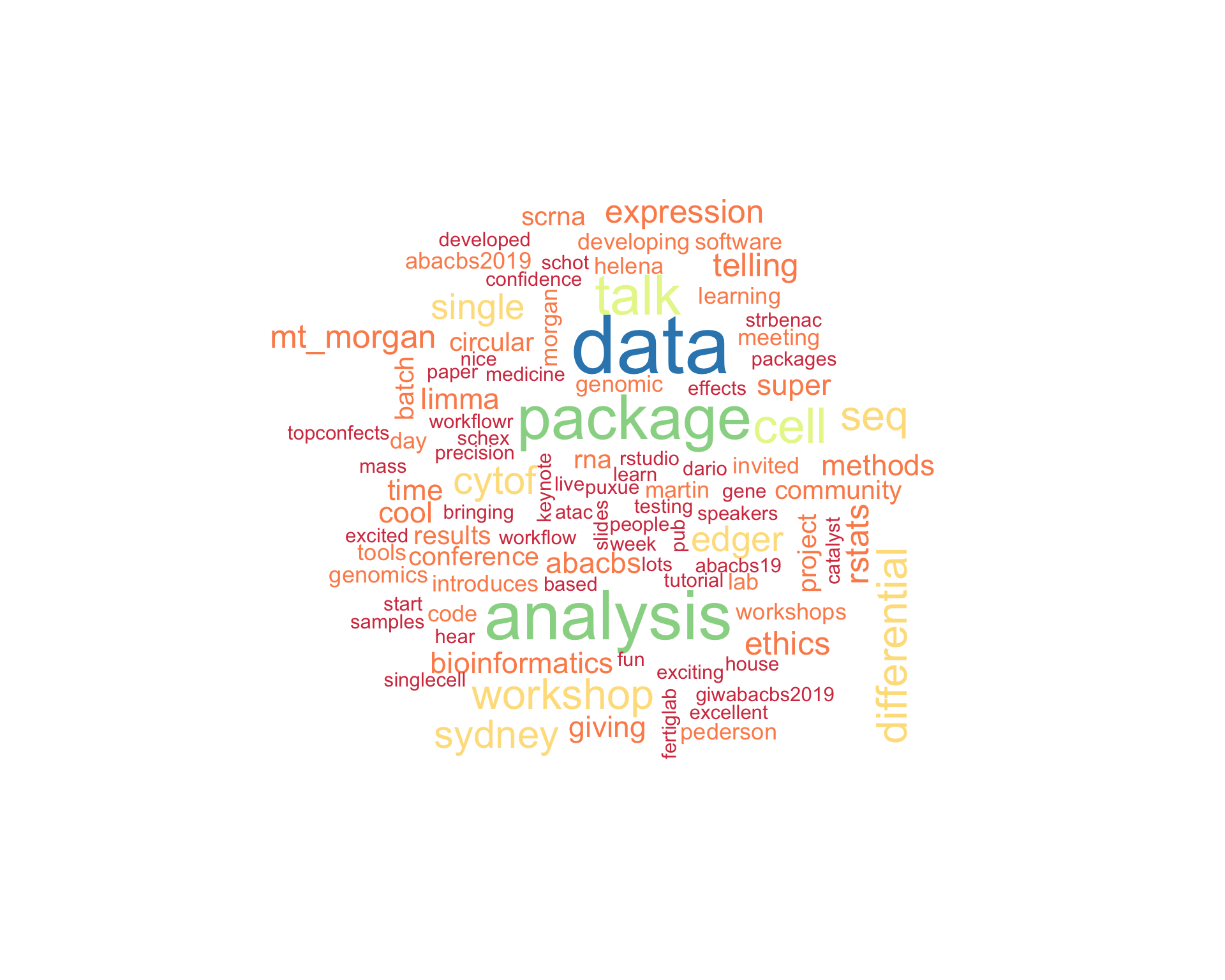
7.1 Word cloud
The top 100 words used 3 or more times.
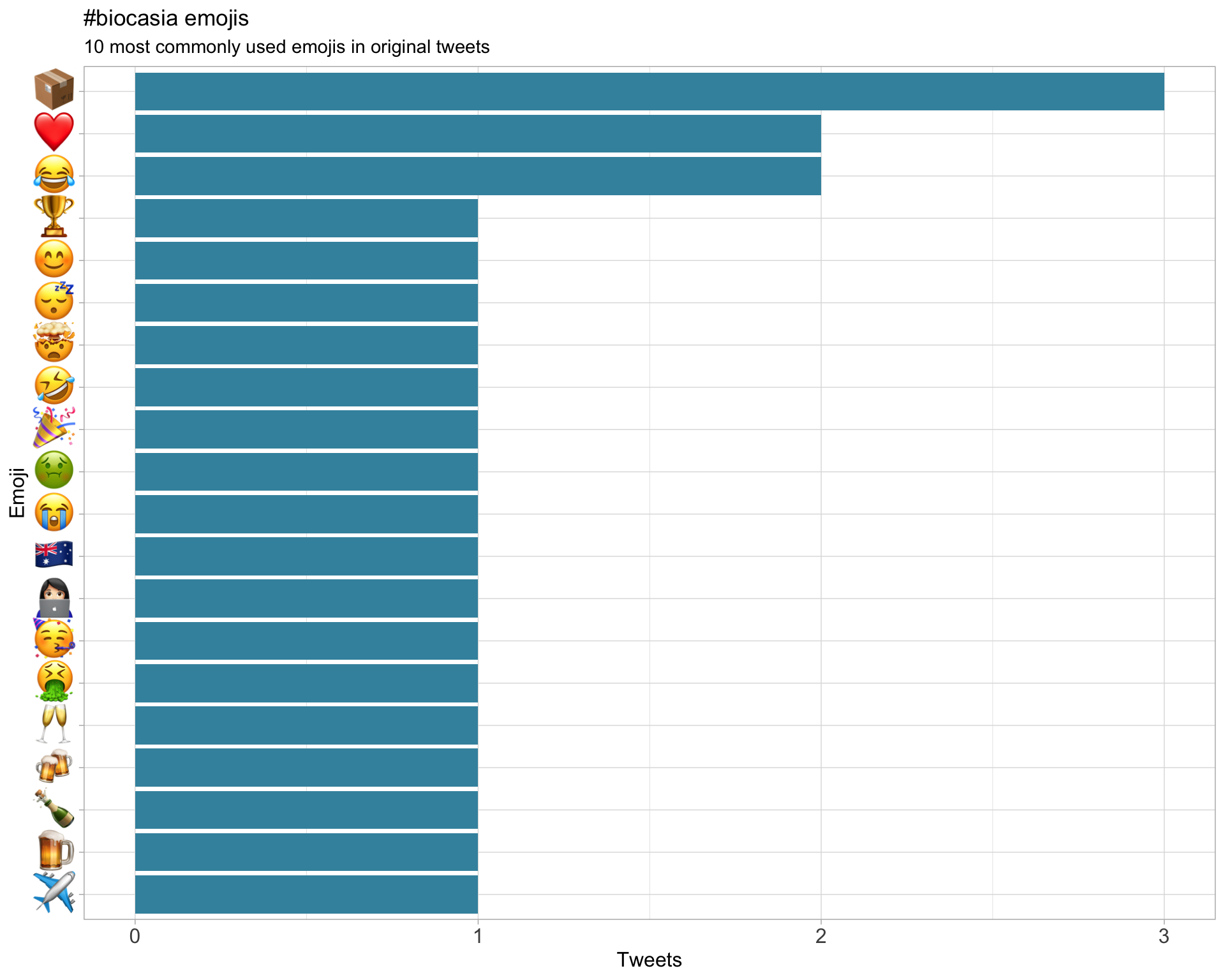
7.3 Emojis
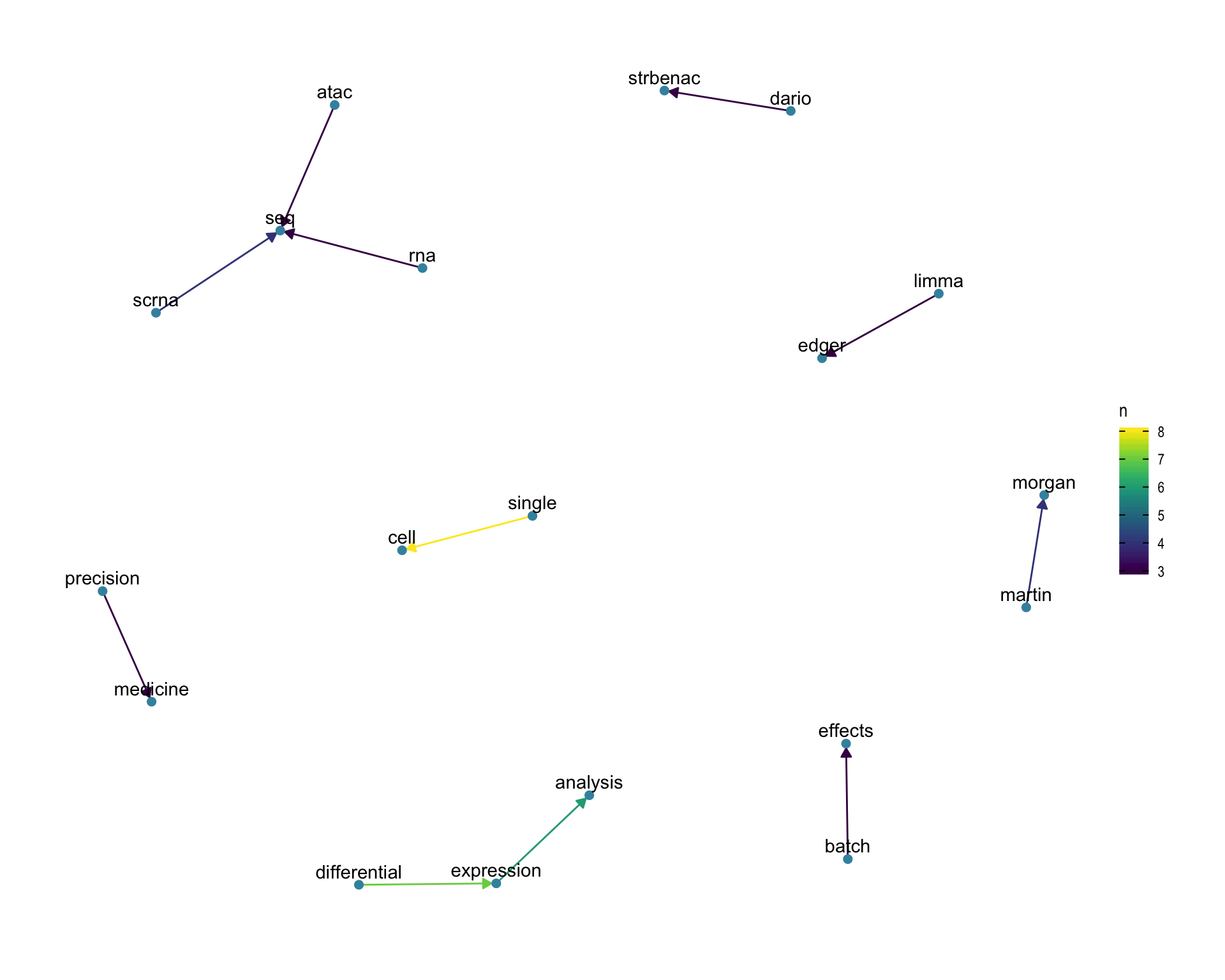
7.4 Bigram graph
Words that were tweeted next to each other at least 3 times.
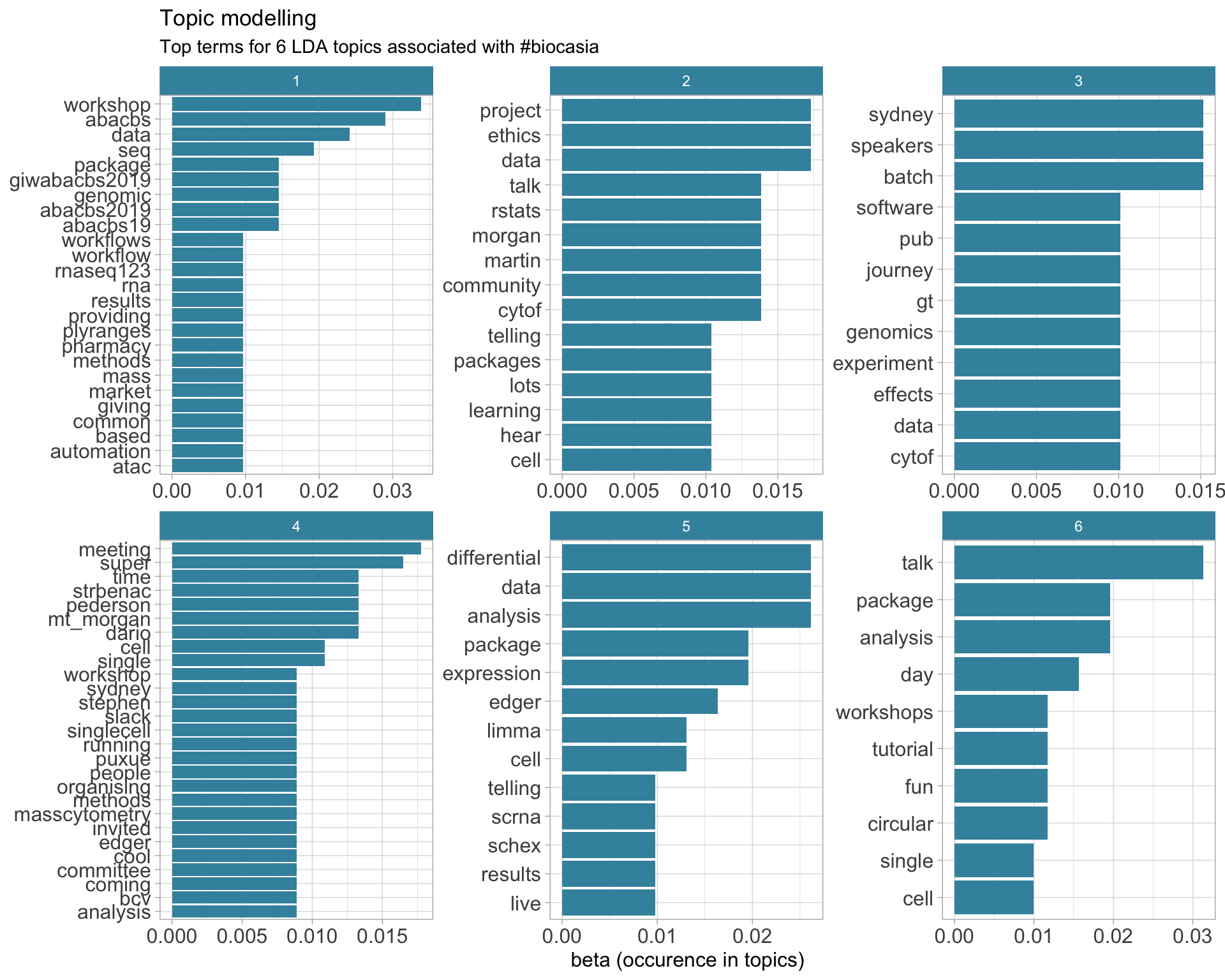
7.5 Topic modelling
Top 10 words associated with 6 topics identified by LDA.
7.5.1 Representative tweets
Most representative tweets for each topic
Topic 1
| screen_name | text | gamma |
|---|---|---|
| paulfharrison |
Workshop by @_StuartLee: plyranges for fluent genomic ranges manipulation, loading Salmon data with tximeta, joining RNA-Seq and ATAC-Seq by range, and a whole lot of useful tricks along the way. #biocasia https://t.co/fnI4sRhwPY |
0.9963609 |
| MnM_HealthCare |
Pharmacy Automation Market: Global Key Players, Trends, Industry Size #Pharmacy #Automation #Market #robot #healthcare #medical #devices #health #healthy #BiocAsia https://t.co/fcBZ9eptTf |
0.9961702 |
| malhamdoosh | Professor Pei Wang from @IcahnMountSinai at #biocasia 2019 explaining her DreamAI method at her workshop on methods and tools for preprocessing and imputation of mass spectrometry-based proteomics data https://t.co/2jZbPIIwaj | 0.9959585 |
| PeteHaitch | There are 27 @Bioconductor workflows for common genomic analyses (https://t.co/7qUnzfWepD) Since we’re at #biocasia/#BioInfoSummer, @mritchieau is highlighting the true blue, dinky di, Aussie-developed RNAseq123 workflow (https://t.co/z5niD5vbBx) available in Chinese and English! | 0.9957220 |
| shazanfar |
For those interested in keeping up to date with the festival of bioinformatics this & next week in Sydney, here’s my Tweetdeck search 😂 #BioInfoSummer OR @bioinfosummer OR #biocasia OR #abacbs OR @abacbs OR #GIWABACBS2019 OR #abacbs2019 OR #abacbs19 did I cover all bases? 😂 |
0.9951549 |
| _StuartLee | For #BiocAsia I’ve written a workflow package for exploring and combining results from RNA-seq and ATAC-seq data. First iteration is up here: https://t.co/03oRjSfThQ | 0.9948110 |
| JovMaksimovic | Using formal data structures and data import functions in @Bioconductor can help you avoid common pitfalls such as dealing with 0 vs. 1 based coordinates #biocasia | 0.9939524 |
| mritchieau | @joshuakho’s paper on this work is at https://t.co/aBQvNFUCcc Great collaboration with David Humphreys from @VictorChangInst driving methods and software development #biocasia https://t.co/OiRtwPzVtg | 0.9939524 |
| trashystats | Last round of workshops. @_StuartLee is giving his very interesting workshop of using plyranges to highlight the genomic context of your DA or DE results. #biocasia https://t.co/fikQxgovdR | 0.9939524 |
| shazanfar |
Added, thanks Andrew!! #BioInfoSummer OR @bioinfosummer OR #biocasia OR #abacbs OR @abacbs OR #GIWABACBS2019 OR #abacbs2019 OR #abacbs19 OR #COMBINE19 OR @combine_au OR #COMBINE2019 https://t.co/GcPXYQY7ML |
0.9934070 |
Topic 2
| screen_name | text | gamma |
|---|---|---|
| shazanfar |
What is ethics? Inner morals & culture play a role, but are not the same as ethical theories. E.g.s: consequentialism (the goodness of the outcome is what matters), deontology (what is moral is good), & virtue ethics (is the person’s motivation good?) #biocasia #bioinfosummer |
0.9959585 |
| PeteHaitch | @Bioconductor invests time and effort into developing reusable data structures that are specifically designed for bioinformatics datasets, like SummarizedExperiment. Using these saves you developing your own and improves interoperability between packages #biocasia https://t.co/WCnMuTI0RK | 0.9959585 |
| annaquagli | @MTrussart discussing all of #Cytof at #biocasia. Chat to her to know more about her debarcoding method using Gaussian mixtures, #reproducibility and removing batch effect with the #ruv #rstats 📦, #normalization and cell #clustering! https://t.co/Ysr9InoDhs https://t.co/2Beaa8jXqU | 0.9959585 |
| lazappi | @shazanfar Yay to #biocasia for having this! 🎉 I think all conferences should have one invited “community talk” that is relevant to the audience but not a research project. Ethics is one example but also patient stories, mental health, workplace practices, career prospects etc. | 0.9959585 |
| PeteHaitch | Delighted to have @CrowellHL as a keynote speaker at #biocasia! Helena and her colleagues in @markrobinsonca’s lab develop great @Bioconductor packages for CyTOF (https://t.co/ifzdnjDbLa) and scRNA-seq (https://t.co/YAYEhwEEus) data. https://t.co/dGh9rwdla7 | 0.9954561 |
| PeteHaitch | Martin Morgan making the pitch at #BioInfoSummer / #biocasia to learn @Bioconductor There’s lots of learning material available: https://t.co/xjRvliJxmb And the project is essential to lots of current research: https://t.co/qppX0pRQTO | 0.9948110 |
| trashystats |
@shazanfar is absolutely filling John’s massive shoes (literally and figuratively) by presenting scHOT which allows investigation of higher order statistics in single cell data #biocasia. https://t.co/FNSAlOjfup |
0.9948110 |
| PeteHaitch | It takes 5+ bioinformaticians to get the bloody projector working, thanks to @JovMaksimovic for being willing to improvise! Excited to hear her work on gene set testing for differentially methylated regions #biocasia https://t.co/R8OUlu4Hy6 | 0.9948110 |
| JovMaksimovic | .@mritchieau telling everyone at #biocasia / #BioInfoSummer about his lab’s RNAseq 123 @Bioconductor workflow and giving a shout out to Melbourne’s local #bioconductor and #rstats Jedi Master - Gordon Smyth. https://t.co/V817fxFEeZ | 0.9948110 |
| mritchieau | @MTrussart introduces us to CyTOF analysis, how to preprocess these data, a novel barcoding strategy to increase cell number and the use of RUV for removing batch effects #biocasia https://t.co/v1tiB4bSlB | 0.9948110 |
Topic 3
| screen_name | text | gamma |
|---|---|---|
| shazanfar | Dr Lisa Drive, bioethicist from the USyd Philosophy dept, discussing concepts of ethics in the context of precision medicine. It’s impossible to deny the wider impact that medical/clinical bioinformatics can have upon members of society #biocasia #bioinfosummer | 0.9965334 |
| PeteHaitch | @MTrussart telling us the best way to deal with batch effects in CyTOF (or indeed any experiment) is to design the experiment properly! If you plan by expecting batch effects, e.g. including control samples in each batch, then you can adjust for them #biocasia https://t.co/DX40j4O1B8 | 0.9963609 |
| carroll_jono | I’ll be in Sydney for #BioCAsia as of tomorrow afternoon - any #rstats peeps willing to brave the smoke for dinner and/or drinks around Circular Quay? I’ve never been to Sydney so I’ll be checking out the gardens, Opera House, and the bridge like a proper tourist. https://t.co/QR8VD62gg4 | 0.9963609 |
| shazanfar | #biocasia @paulfharrison presenting Topconfects (pub https://t.co/5mYDprcT27 ) that ranks genes based on confidence bounds on the log-fold changes. Furthering @davisjmcc & Smyth’s TREAT (pub https://t.co/iZeszEIkDU ) Software: https://t.co/5H3KA4lFZs works nicely with limma/edgeR | 0.9961702 |
| shazanfar |
#BioInfoSummer #biocasia @FertigLab describes software for performing non-negative matrix factorisation (CoGAPS) and projection transfer learning approaches (projectR) for high throughput genomics data https://t.co/5FojtTftWe |
0.9959585 |
| Al__Forrest | Enjoying #biocasia We have a great lineup of speakers in human genomics at #HGM2020 in Perth April 5-8 https://t.co/S4dGO1LRpe Earlybird closes Dec 16th. Over 40 speakers to be selected from the submitted abstracts!! | 0.9957220 |
| annaquagli | Thanks to @trashystats for the detailed documentation and workshop on making @Bioconductor 📦 . Did you know that you can go inside the R/myfun.R -> click on ‘Code’ -> ‘Insert Roxygen Skeleton’ ?🤯 #BiocAsia https://t.co/GDDjQW9Gny https://t.co/urkJfLMP7v | 0.9948110 |
| mritchieau | And that’s a wrap for #BioInfoSummer #biocasia 2019. Congrats to the poster prize winners and thanks again to all the speakers, participants, organisers and generous sponsors especially @DiscoverAMSI for an awesome event! #staycurious https://t.co/e4JvGiYzKm | 0.9948110 |
| PeteHaitch | @rafalab presenting his work estimating death rates following Hurricane Maria in Puerto Rico. Here’s the blog post showing how much of the work was in cleaning the data https://t.co/gi8ixEkSZQ #biocasia #BioInfoSummer | 0.9944145 |
| PNgsabrina | Beautiful work and personal journey by @CrowellHL from @UZH_en on the masterminds behind CyTOF, Catalyst, then muscat. #bioconductor #BiocAsia @Bioinfosummer 😊 @DiscoverAMSI It’s inspiring to see one the cherishs her work with great passion. https://t.co/tI9wHl4jbK | 0.9939524 |
Topic 4
| screen_name | text | gamma |
|---|---|---|
| annaquagli | Super interesting discussion by Charity Law from @mritchieau lab about how different levels of variation (BCV in the #edgeR world) infleluence the significance of differential expression analysis… PS. I have rarely seen BCV < 0.4/0.5 in my samples 😭 #biocasia https://t.co/NlXzghbNb7 | 0.9968337 |
| shazanfar |
PuXue Xiao with her newly minted PhD (congrats!!), postdoc with @davisjmcc presenting a great talk on the important problem of nearest-neighbour graph construction prior to single cell clustering #biocasia PuXue will be presenting a single cell workshop later today! https://t.co/7YOssXZpGN |
0.9965334 |
| PeteHaitch | The “boring to some people but I think is really cool result” (@CrowellHL’s words, but I concur) is that running edgeR or limma on the summed counts works best and is many, many times faster and simpler than running a full-blown mixed model in their experience #biocasia https://t.co/YdYzI7UnpJ | 0.9965334 |
| mritchieau | @mt_morgan encourages us to participate in #bioconductor #slack. Has been a great vehicle for community participation & collaboration. Highlights include development of iSEE & the #singlecell analysis cookbook https://t.co/CLrXYW2kmB. Sign up at https://t.co/Qa7OwOPQdG #biocasia https://t.co/tRJlxAPWYY | 0.9961702 |
| DrHelenMcGuire | #IAmAnImmunologist is INVITED SPEAKING! Super pleased to be invited to share my passion for #immunology and #highdimensional #masscytometry at #Bioinfosummer #biocasia @Bioinfosummer hosted at @Sydney_Uni @CPC_EMCR @SydneyCytometry @ASImmunology @ACScytometry https://t.co/Ln14hNjSyM | 0.9959585 |
| shazanfar | #biocasia 2019 is now coming to a close, organiser @PeteHaitch closing the meeting with news that BiocAsia will be coming to China in 2020! Thank you to the organising committee Pete, Steve Pederson, @trashystats and Dario Strbenac for an excellent meeting! https://t.co/z26y8LFOnI | 0.9959585 |
| JovMaksimovic | Cab driver got me to Sydney airport in plenty of time for my flight, despite peak hour traffic. It was the jerkiest ride though - my stomach is still recovering from the g-forces 🤢🤮 #biocasia | 0.9951549 |
| JovMaksimovic | I’ve a great time at #biocasia but it’s nearly time to jet back to Melbourne. Thanks to the organising committee for a fab meeting: @PeteHaitch @trashystats Dario Strbenac and Stephen Pederson 🏆 | 0.9939524 |
| mritchieau | So many cool papers @FertigLab @GenesOfEve and colleagues to learn more about these methods e.g. https://t.co/OsJH622gAo and https://t.co/woblN8CxhC #biocasia https://t.co/7NhkR0zyuG | 0.9934070 |
| brian_gloss | hey #Bioinfosummer #biocasia people (talking about spillover), is it possible to just consider each fluorophore from its distribution across n-dimensions (depending on detectors) and do away with correcting? | 0.9927534 |
| brian_gloss | @CrowellH highlighting the vital importance of a ground truth in evaluating methods especially in emerging technologies #Bioinfosummer #biocasia | 0.9927534 |
Topic 5
| screen_name | text | gamma |
|---|---|---|
| mritchieau | @paulfharrison introduces us to the ‘False Coverage-statement Rate’ concept which seems like a neat idea. Works with limma, edgeR, DESeq2 differential expression analysis pipelines and can be used in #geneset testing. Slides on topconfects at https://t.co/wbAOqjtCw2 #biocasia https://t.co/tHtpyYPWg9 | 0.9965334 |
| shazanfar | #bioinfosummer #biocasia Elana Fertig @FertigLab presenting work on non-negative matrix factorisation techniques for genomics data. Using the gamma distribution (equiv to sum of exponentials) allows for generalisability as well as reflecting sparsity in the underlying system https://t.co/ZCKtjN8743 | 0.9963609 |
| PeteHaitch | Charity Law, developer of voom, introducing an extension to voom to handle group-specific variance estimation and how she’s thinking about adapting it to scRNA-seq differential expression analysis #biocasia https://t.co/HOguf3wWpl | 0.9961702 |
| PeteHaitch | By constructing ‘psuedobulk’ samples from scRNA-seq data, we can use tried and tested methods for differential expression analysis (limma, edgeR, DESeq2, etc) @CrowellHL benchmarked this approach along with other methods in https://t.co/SMNPfC7iyO #biocasia https://t.co/jZwcPFN13i | 0.9961702 |
| CFlensburg |
FOMO alert! Everyone in bioinformatics and #rstats should have a scroll through the bioconductor #biocasia hashtag from the last couple of days! And Monday next week I’m joining Sydney for the bioinformatics conference #abacbs2019, I might be live-tweeting a bit. :) https://t.co/scPHdyoNcu |
0.9961702 |
| PeteHaitch | We’ve had scRNA, CyTOF, and now Hi-C with @HannahCoughlan4 telling us about a gene-oriented analysis approach using limma and edgeR, tools originally developed for differential expression analysis. Absolute workhorses of @Bioconductor #biocasia https://t.co/4ZI4qf6rkD | 0.9959585 |
| mritchieau | @DongXueyi tells us how to analyse #longread @nanopore #RNAseq data using #bioconductor. Schmd1 data (both long and short reads) from @BlewittMarnie lab. New tools still needed for accurate isoform analysis #biocasia https://t.co/rjrExr6jv6 | 0.9957220 |
| PeteHaitch | @paulfharrison advocating for the use of confidence intervals over p-values when reporting differential expression analysis results #biocasia This idea implemented in topconfects @Bioconductor package: https://t.co/8bKQQk0Bg2 Slides: https://t.co/qJqFLwWzbB https://t.co/O3qL6A7Ov7 | 0.9957220 |
| shazanfar | #biocasia #bioinfosummer @trashystats addressing the issue of overplotting of many many coloured points in a scatterplot, with Bioconductor package schex (pron “ess-see-hex” 🤣). Saskia is super cool under the live demo pressure, well done!! https://t.co/mywX9y9MMN | 0.9957220 |
| PeteHaitch | Closing out the first session of #biocasia, we’ve got our first presentation from an undergrad! Kathleen Zeglinski telling us about gmoviz, a package she is developing at @CSL to visualise genome-editing data https://t.co/GsixCMALXD | 0.9954561 |
Topic 6
| screen_name | text | gamma |
|---|---|---|
| PeteHaitch | Steve Pederson breaking the space-time continuum in 5min talk at #biocasia, reading out a letter to his future self from his past self about the experience of submitting his first @Bioconductor package. Love someone who gets creative for a presentation https://t.co/XfcLxmbZYW | 0.9961702 |
| CrowellHL | Off to 🇦🇺 for some Bioinformatics fun 👩🏻💻, to talk & teach about CyTOF at #BiocAsia / BioInfoSummer in Sydney, and explore other parts of AUS. Stressed, grateful and excited! - Now looking forward to 20h of reading & movies ✈️😴 That completes my #BiocBingo 🥳 | 0.9957220 |
| shazanfar | Phenomenal start to Helena Crowell’s @CrowellHL ( @markrobinsonca lab ) #biocasia #bioinfosummer keynote on Differential discovery in high-dimensional cytometry data. Beginning with a timeline of important work in this space by a number of key contributors https://t.co/VStuFvtEMA | 0.9957220 |
| mritchieau | @shazanfar from the @MarioniLab introduces scHOT for flexible analysis of higher-order interactions in #singlecell data: shows us examples of variability, correlation and spatial analysis #biocasia. Code available at https://t.co/p8l2m21jqt https://t.co/41Nbf73QtU | 0.9954561 |
| shazanfar | #biocasia Joshua Ho @joshuawkho presenting Ularcirc (a back-splice event on Circular :D ), circular RNA analysis package available on @Bioconductor https://t.co/jFxiHNhnOg Tutorial video https://t.co/bnZhk3tNCu | 0.9948110 |
| PeteHaitch | Exciting talk by @shazanfar bringing scHOT (https://t.co/1NSzfqWwLe) to spatial transcriptomics, the cool new kid on the single cell block #biocasia https://t.co/4SsGLInsmk | 0.9944145 |
| DiscoverAMSI | The first of the AMSI #BioInfoSummer 2019 joint sessions with #biocasia will start at 1.30pm with 3 workshops to choose from sponsored by @CMRI_AUS on our mass spec analytics day | 0.9944145 |
| PeteHaitch | PuXue Qiao sharing the resources she, along with Ruqian Lyu and @davisjmcc, developed for a 2-day scRNA-seq course https://t.co/sq2q2HgEA6 #biocasia | 0.9934070 |
| shazanfar |
Great talk by Ruqian Lyu @Ruqian_lyu on calculating genetic length from crossover events using COmapR, on Bioconductor! https://t.co/VNTMQBJAj1 #BioInfoSummer #biocasia |
0.9934070 |
| mritchieau | Watch the demo of using the Ularcirc #bioconductor package for visualising and analysing circular RNAs at https://t.co/P0ViJ7pi43 #biocasia https://t.co/3olI9ltMST | 0.9927534 |
| One_News_Page | Afghans demand justice amid attempt to probe war crimes at ICC: https://t.co/atyUQAyww8 #biocasia | 0.9927534 |
| PeteHaitch | @haoyyang bringing us home on Day 1 with an overview of software for single-cell ATAC-seq #biocasia https://t.co/RyAq7Zhyhz | 0.9927534 |
8 Links
Links to GitHub, GitLab, BitBucket, Bioconductor or CRAN mentioned in Tweets.
| Name | Tweets | Retweets | Type | Link |
|---|---|---|---|---|
| CATALYST | 1 | 0 | Bioc | bioconductor.org/packages/CATALYST |
| CoGAPS.html | 1 | 0 | Bioc | bioconductor.org/packages/CoGAPS.html |
| muscat | 1 | 0 | Bioc | bioconductor.org/packages/muscat |
| projectR.html | 1 | 0 | Bioc | bioconductor.org/packages/projectR.html |
| scHOT2019 | 1 | 0 | GitHub | marionilab/schot2019 |
| topconfects | 1 | 0 | Bioc | bioconductor.org/packages/topconfects |
| topconfects.html | 1 | 0 | Bioc | bioconductor.org/packages/topconfects.html |
| ttBulk | 1 | 0 | GitHub | stemangiola/ttbulk |
| Ularcirc.html | 1 | 0 | Bioc | bioconductor.org/packages/Ularcirc.html |
| muscat-comparison | NA | NA | GitHub | helenalc/muscat-comparison/blob/master/magl/docs/index.html |
Session info
## ─ Session info ───────────────────────────────────────────────────────────────
## setting value
## version R version 4.0.0 (2020-04-24)
## os macOS Catalina 10.15.6
## system x86_64, darwin17.0
## ui X11
## language (EN)
## collate en_US.UTF-8
## ctype en_US.UTF-8
## tz Europe/Berlin
## date 2020-09-01
##
## ─ Packages ───────────────────────────────────────────────────────────────────
## package * version date lib source
## askpass 1.1 2019-01-13 [1] CRAN (R 4.0.0)
## assertthat 0.2.1 2019-03-21 [1] CRAN (R 4.0.0)
## backports 1.1.8 2020-06-17 [1] CRAN (R 4.0.0)
## bitops 1.0-6 2013-08-17 [1] CRAN (R 4.0.0)
## callr 3.4.3 2020-03-28 [1] CRAN (R 4.0.0)
## clamour * 0.1.0 2020-09-01 [1] Github (lazappi/clamour@c8ea1c7)
## cli 2.0.2 2020-02-28 [1] CRAN (R 4.0.0)
## colorspace 1.4-1 2019-03-18 [1] CRAN (R 4.0.0)
## crayon 1.3.4 2017-09-16 [1] CRAN (R 4.0.0)
## curl 4.3 2019-12-02 [1] CRAN (R 4.0.0)
## digest 0.6.25 2020-02-23 [1] CRAN (R 4.0.0)
## dplyr * 1.0.1 2020-07-31 [1] CRAN (R 4.0.2)
## ellipsis 0.3.1 2020-05-15 [1] CRAN (R 4.0.0)
## emo * 0.0.0.9000 2020-08-17 [1] Github (hadley/emo@3f03b11)
## evaluate 0.14 2019-05-28 [1] CRAN (R 4.0.0)
## fansi 0.4.1 2020-01-08 [1] CRAN (R 4.0.0)
## farver 2.0.3 2020-01-16 [1] CRAN (R 4.0.0)
## forcats * 0.5.0 2020-03-01 [1] CRAN (R 4.0.0)
## fs * 1.5.0 2020-07-31 [1] CRAN (R 4.0.2)
## generics 0.0.2 2018-11-29 [1] CRAN (R 4.0.0)
## ggforce 0.3.2 2020-06-23 [1] CRAN (R 4.0.2)
## ggplot2 * 3.3.2 2020-06-19 [1] CRAN (R 4.0.2)
## ggraph * 2.0.3 2020-05-20 [1] CRAN (R 4.0.0)
## ggrepel * 0.8.2 2020-03-08 [1] CRAN (R 4.0.0)
## ggtext * 0.1.0 2020-06-04 [1] CRAN (R 4.0.2)
## glue 1.4.1 2020-05-13 [1] CRAN (R 4.0.0)
## graphlayouts 0.7.0 2020-04-25 [1] CRAN (R 4.0.0)
## gridExtra 2.3 2017-09-09 [1] CRAN (R 4.0.0)
## gridtext 0.1.1 2020-02-24 [1] CRAN (R 4.0.2)
## gtable 0.3.0 2019-03-25 [1] CRAN (R 4.0.0)
## here * 0.1 2017-05-28 [1] CRAN (R 4.0.0)
## highr 0.8 2019-03-20 [1] CRAN (R 4.0.0)
## htmltools 0.5.0 2020-06-16 [1] CRAN (R 4.0.0)
## httr 1.4.2 2020-07-20 [1] CRAN (R 4.0.2)
## igraph * 1.2.5 2020-03-19 [1] CRAN (R 4.0.0)
## janeaustenr 0.1.5 2017-06-10 [1] CRAN (R 4.0.0)
## jsonlite 1.7.0 2020-06-25 [1] CRAN (R 4.0.0)
## kableExtra * 1.2.1 2020-08-27 [1] CRAN (R 4.0.2)
## knitr * 1.29 2020-06-23 [1] CRAN (R 4.0.0)
## labeling 0.3 2014-08-23 [1] CRAN (R 4.0.0)
## lattice 0.20-41 2020-04-02 [1] CRAN (R 4.0.0)
## lifecycle 0.2.0 2020-03-06 [1] CRAN (R 4.0.0)
## lubridate * 1.7.9 2020-06-08 [1] CRAN (R 4.0.0)
## magick * 2.4.0 2020-06-23 [1] CRAN (R 4.0.0)
## magrittr 1.5 2014-11-22 [1] CRAN (R 4.0.0)
## markdown 1.1 2019-08-07 [1] CRAN (R 4.0.0)
## MASS 7.3-51.6 2020-04-26 [1] CRAN (R 4.0.0)
## Matrix 1.2-18 2019-11-27 [1] CRAN (R 4.0.0)
## modeltools 0.2-23 2020-03-05 [1] CRAN (R 4.0.0)
## munsell 0.5.0 2018-06-12 [1] CRAN (R 4.0.0)
## NLP 0.2-0 2018-10-18 [1] CRAN (R 4.0.0)
## openssl 1.4.2 2020-06-27 [1] CRAN (R 4.0.0)
## pillar 1.4.6 2020-07-10 [1] CRAN (R 4.0.2)
## pkgconfig 2.0.3 2019-09-22 [1] CRAN (R 4.0.0)
## plyr 1.8.6 2020-03-03 [1] CRAN (R 4.0.0)
## png 0.1-7 2013-12-03 [1] CRAN (R 4.0.0)
## polyclip 1.10-0 2019-03-14 [1] CRAN (R 4.0.0)
## processx 3.4.3 2020-07-05 [1] CRAN (R 4.0.2)
## ps 1.3.3 2020-05-08 [1] CRAN (R 4.0.0)
## purrr * 0.3.4 2020-04-17 [1] CRAN (R 4.0.0)
## R6 2.4.1 2019-11-12 [1] CRAN (R 4.0.0)
## RColorBrewer * 1.1-2 2014-12-07 [1] CRAN (R 4.0.0)
## Rcpp 1.0.5 2020-07-06 [1] CRAN (R 4.0.0)
## RCurl 1.98-1.2 2020-04-18 [1] CRAN (R 4.0.0)
## reshape2 1.4.4 2020-04-09 [1] CRAN (R 4.0.0)
## rlang 0.4.7 2020-07-09 [1] CRAN (R 4.0.2)
## rmarkdown 2.3 2020-06-18 [1] CRAN (R 4.0.0)
## rprojroot 1.3-2 2018-01-03 [1] CRAN (R 4.0.0)
## rstudioapi 0.11 2020-02-07 [1] CRAN (R 4.0.0)
## rtweet * 0.7.0 2020-01-08 [1] CRAN (R 4.0.0)
## rvest * 0.3.6 2020-07-25 [1] CRAN (R 4.0.2)
## scales 1.1.1 2020-05-11 [1] CRAN (R 4.0.0)
## selectr 0.4-2 2019-11-20 [1] CRAN (R 4.0.0)
## sessioninfo 1.1.1 2018-11-05 [1] CRAN (R 4.0.0)
## slam 0.1-47 2019-12-21 [1] CRAN (R 4.0.0)
## SnowballC 0.7.0 2020-04-01 [1] CRAN (R 4.0.0)
## stringi 1.4.6 2020-02-17 [1] CRAN (R 4.0.0)
## stringr * 1.4.0 2019-02-10 [1] CRAN (R 4.0.0)
## tibble 3.0.3 2020-07-10 [1] CRAN (R 4.0.2)
## tidygraph 1.2.0 2020-05-12 [1] CRAN (R 4.0.0)
## tidyr * 1.1.1 2020-07-31 [1] CRAN (R 4.0.2)
## tidyselect 1.1.0 2020-05-11 [1] CRAN (R 4.0.0)
## tidytext * 0.2.5 2020-07-11 [1] CRAN (R 4.0.2)
## tm 0.7-7 2019-12-12 [1] CRAN (R 4.0.0)
## tokenizers 0.2.1 2018-03-29 [1] CRAN (R 4.0.0)
## topicmodels * 0.2-11 2020-04-19 [1] CRAN (R 4.0.0)
## tweenr 1.0.1 2018-12-14 [1] CRAN (R 4.0.0)
## usethis 1.6.1 2020-04-29 [1] CRAN (R 4.0.0)
## utf8 1.1.4 2018-05-24 [1] CRAN (R 4.0.0)
## vctrs 0.3.2 2020-07-15 [1] CRAN (R 4.0.2)
## viridis * 0.5.1 2018-03-29 [1] CRAN (R 4.0.0)
## viridisLite * 0.3.0 2018-02-01 [1] CRAN (R 4.0.0)
## webshot * 0.5.2 2019-11-22 [1] CRAN (R 4.0.0)
## withr 2.2.0 2020-04-20 [1] CRAN (R 4.0.0)
## wordcloud * 2.6 2018-08-24 [1] CRAN (R 4.0.0)
## xfun 0.16 2020-07-24 [1] CRAN (R 4.0.2)
## xml2 * 1.3.2 2020-04-23 [1] CRAN (R 4.0.0)
## yaml 2.2.1 2020-02-01 [1] CRAN (R 4.0.0)
##
## [1] /Library/Frameworks/R.framework/Versions/4.0/Resources/library