ABACBSGIW19
ABACBS/GIW conference 2019
Last built: 2020-09-01 17:53:40
| Parameter | Value |
|---|---|
| hashtag | #ABACBSGIW19 |
| start_day | 2019-12-09 |
| end_day | 2019-12-11 |
| timezone | Australia/Sydney |
| theme | theme_light |
| accent | #de3b30 |
| accent2 | #EE9D97 |
| kcore | 2 |
| topics_k | 6 |
| bigram_filter | 3 |
| fixed | TRUE |
| seed | 1 |
Introduction
An analysis of tweets from the #ABACBSGIW19 hashtag for the joint ABACBS/GIW conference 2019.
A total of 1133 tweets from 344 users were collected using the rtweet R package.
1 Timeline
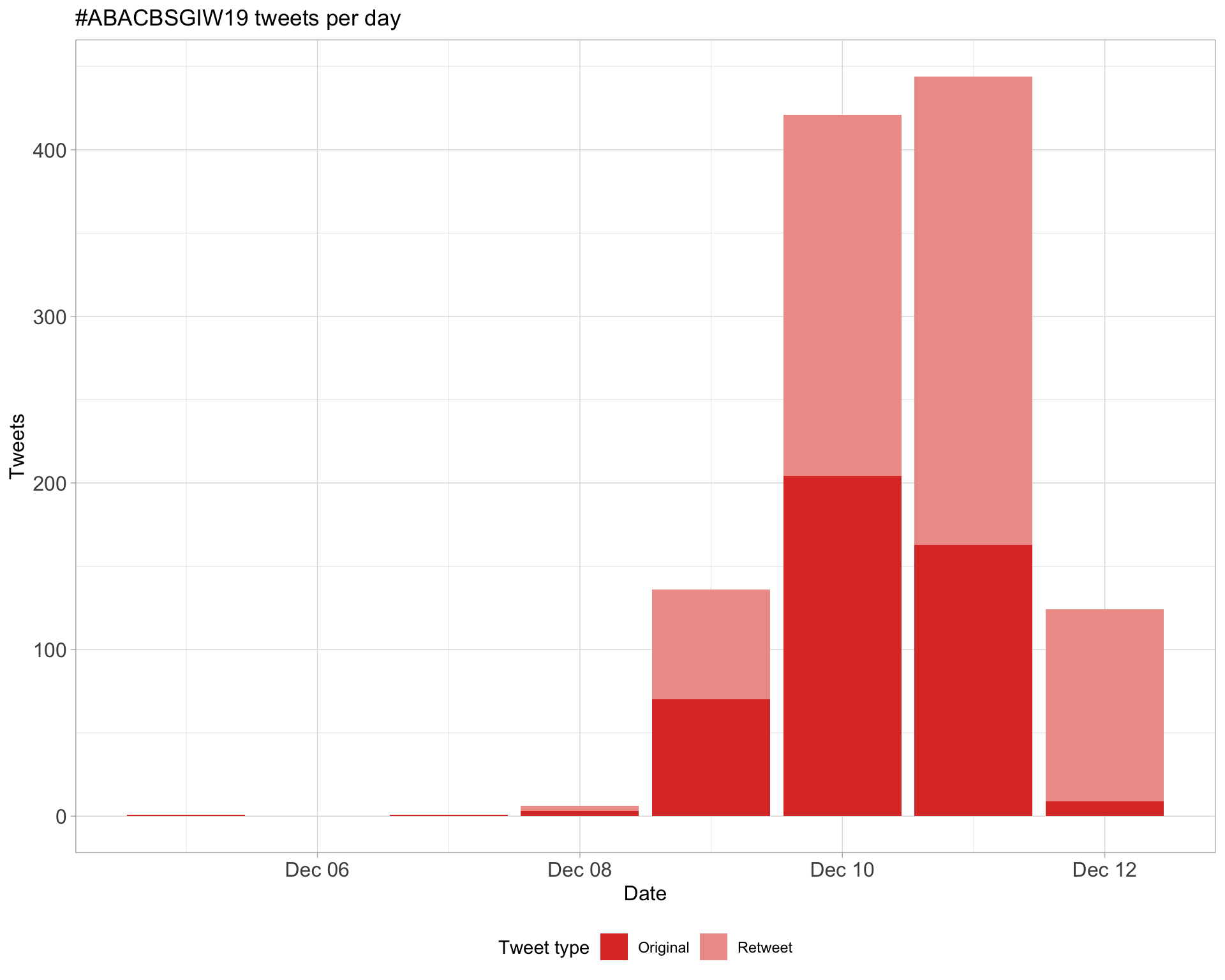
1.1 Tweets by day
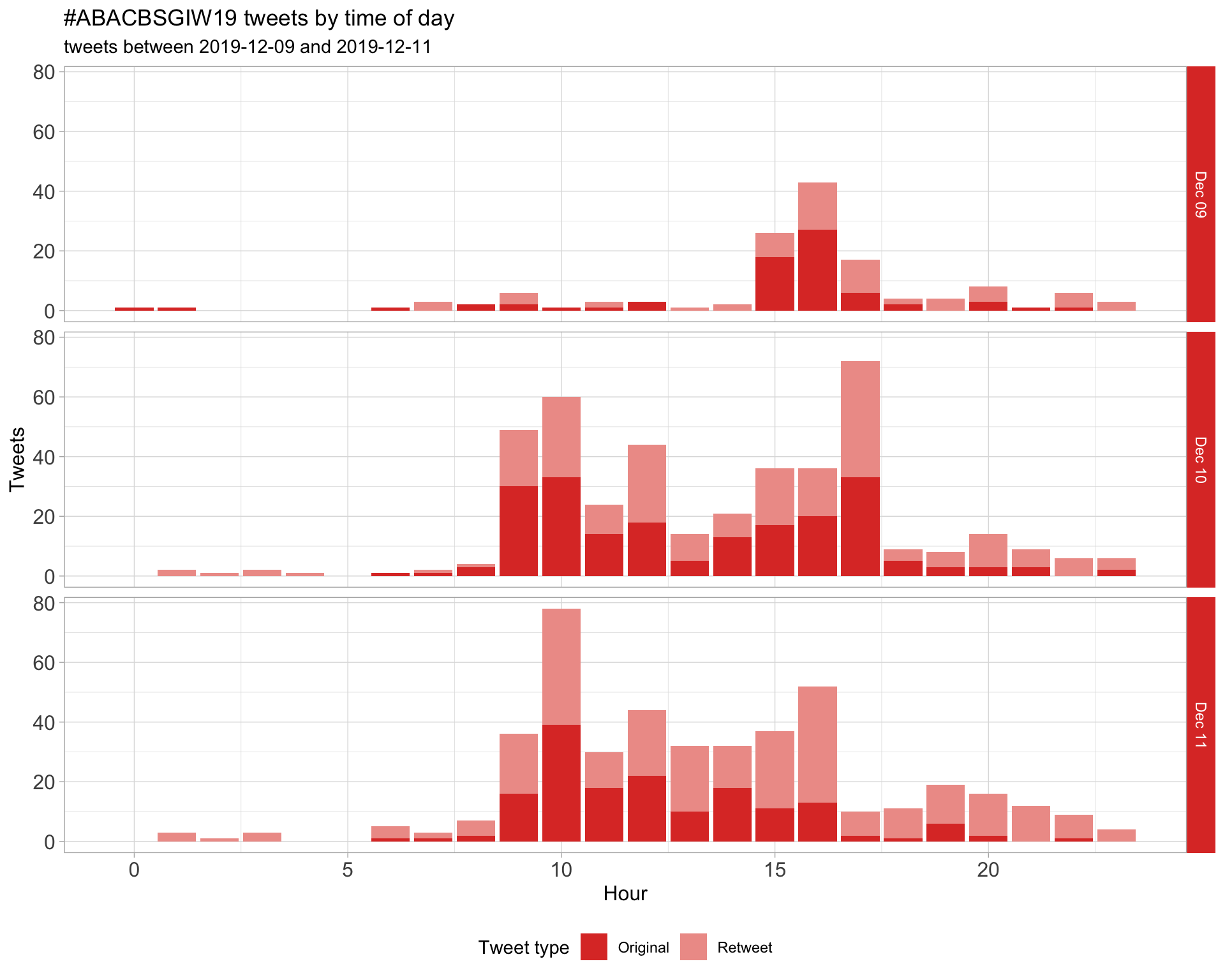
1.2 Tweets by day and time
Filtered for dates 2019-12-09 - 2019-12-11 in the Australia/Sydney timezone.
2 Users
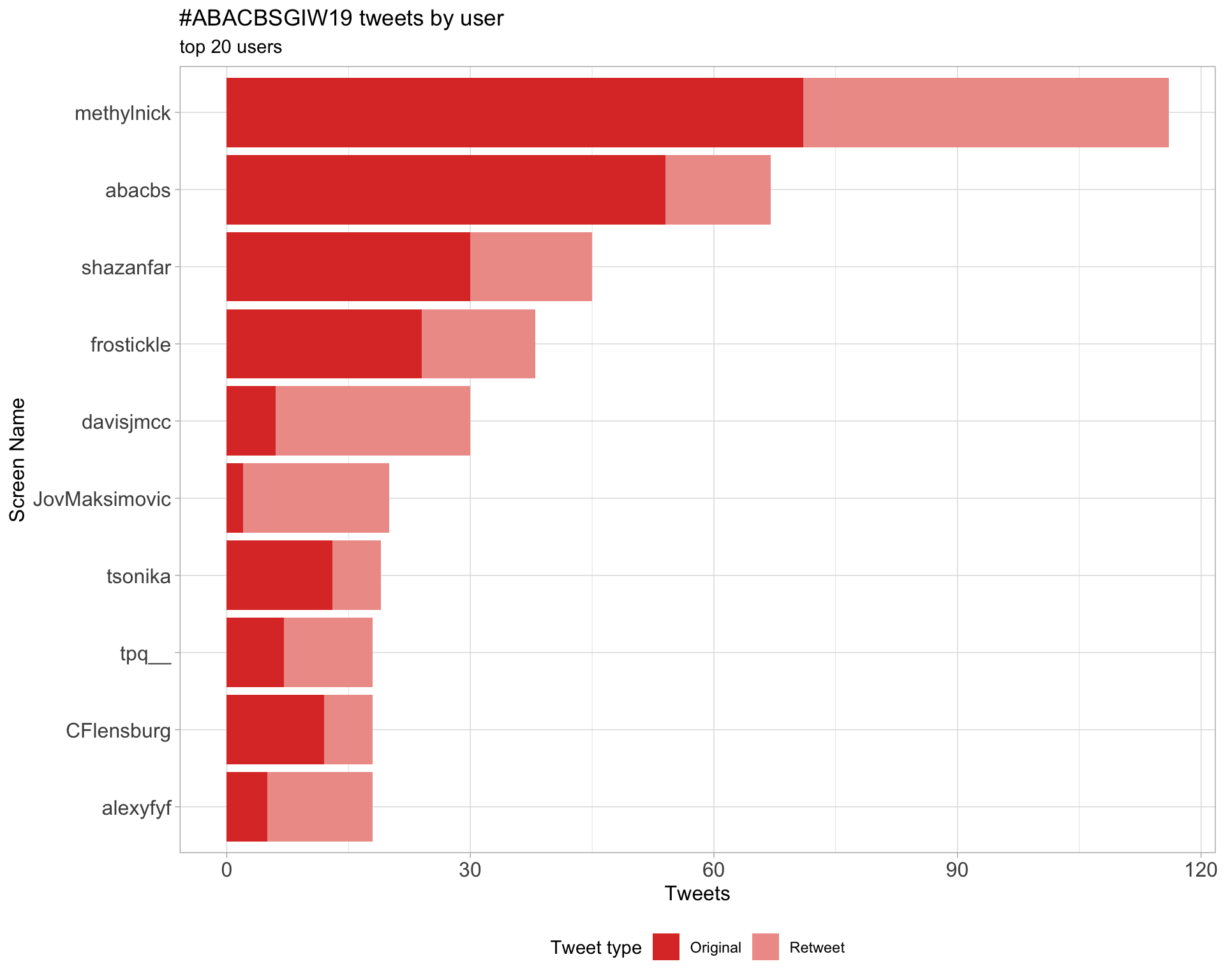
2.1 Top tweeters
Overall
Original
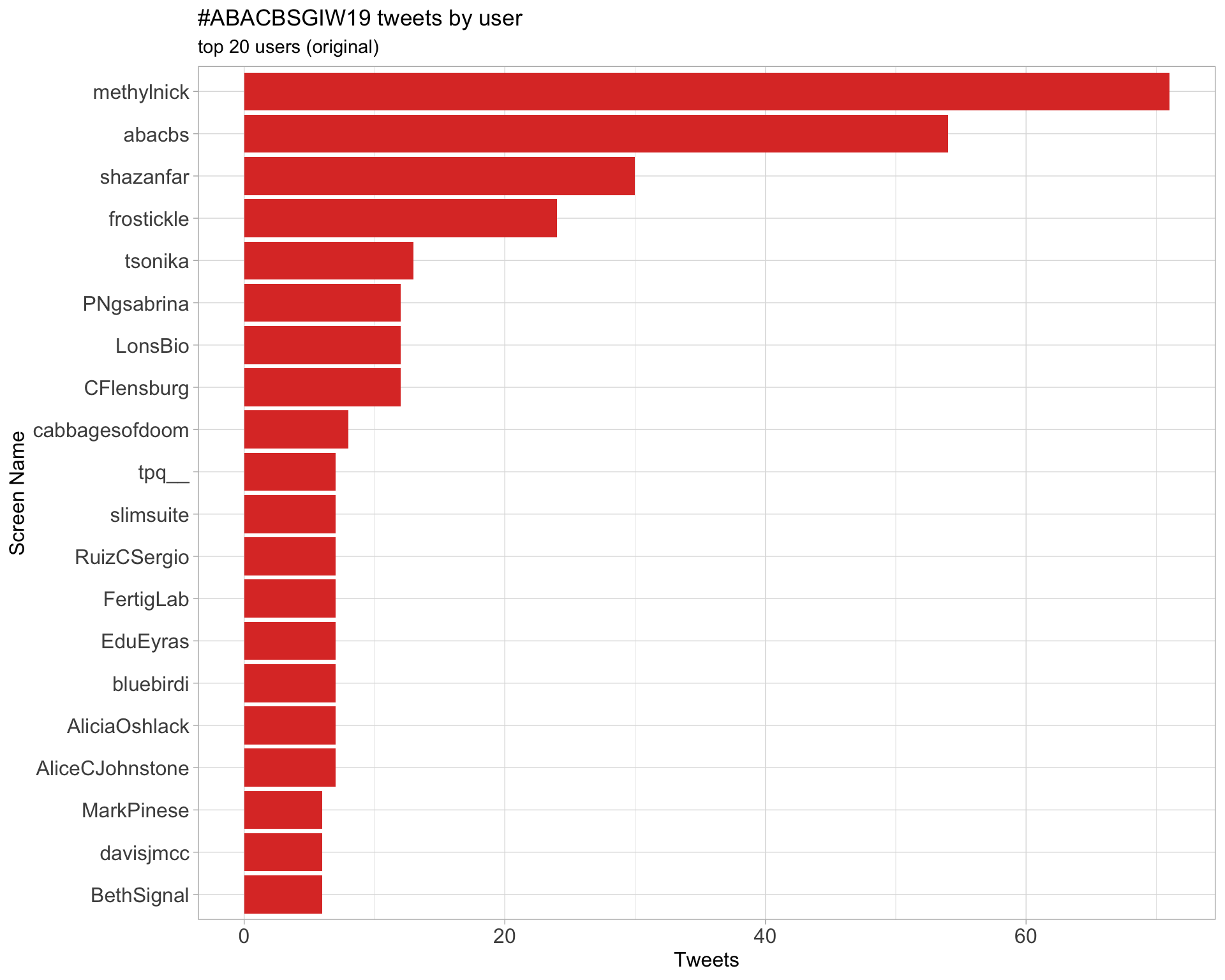
Retweets
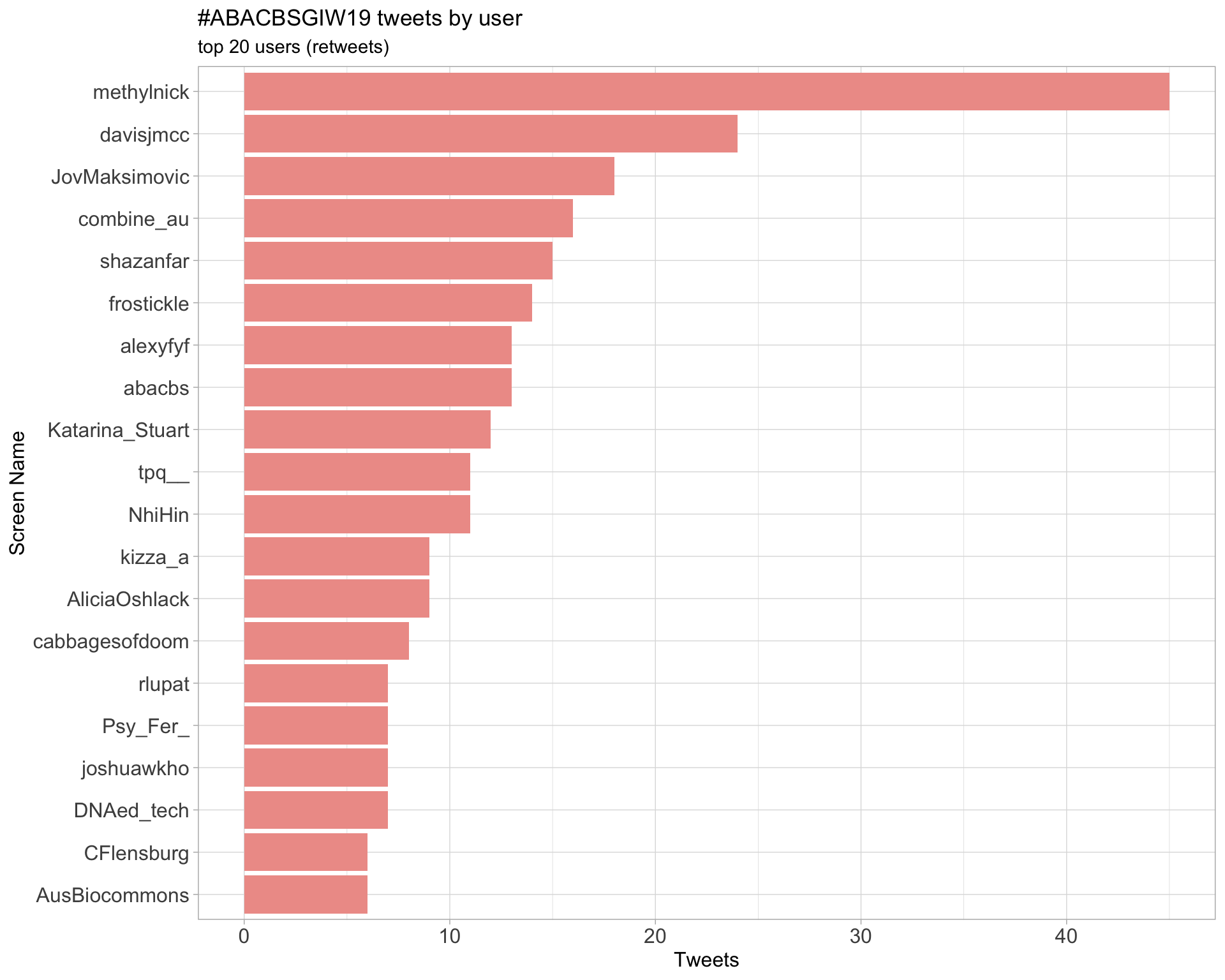
2.2 Retweet proportion
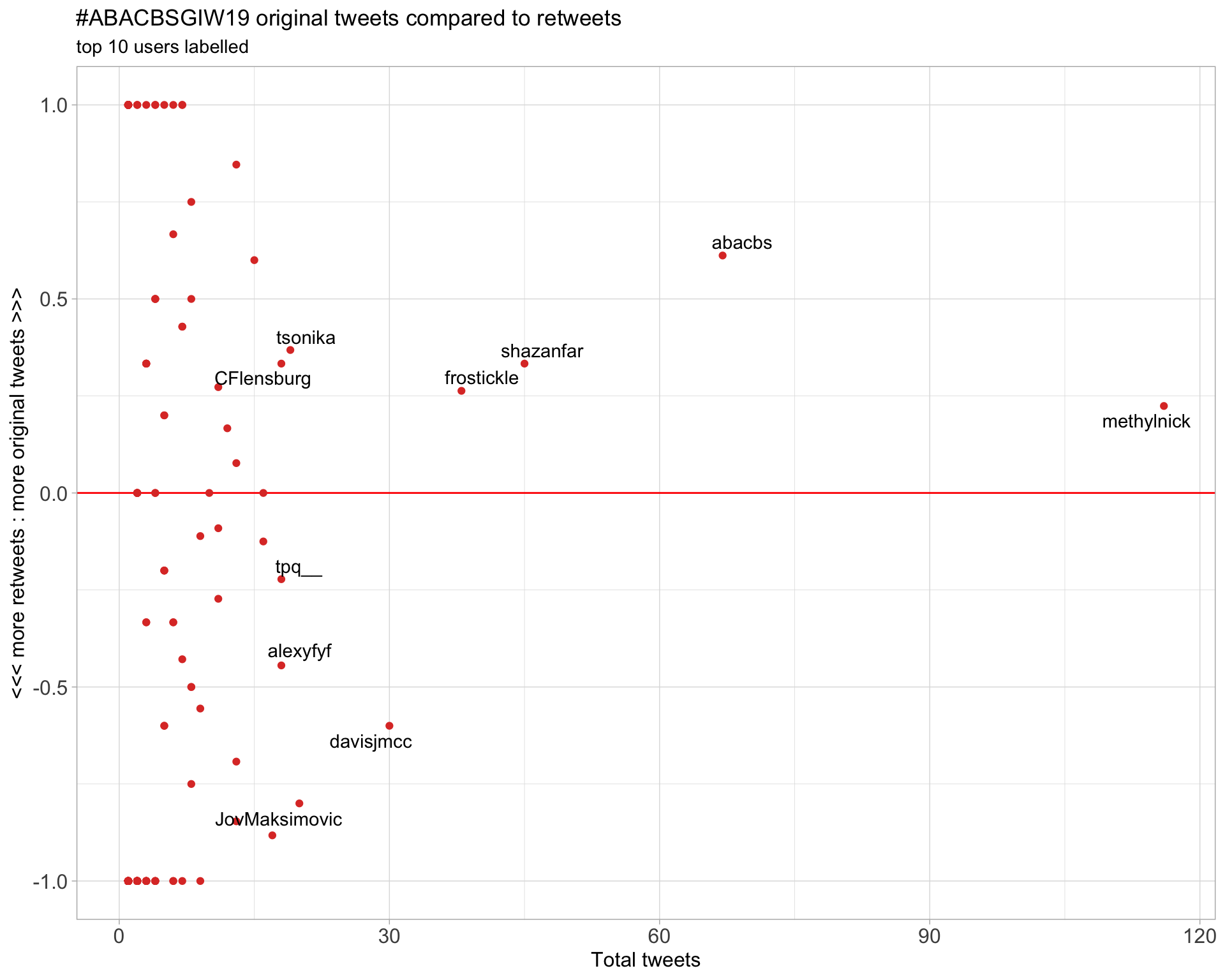
2.3 Top tweeters timeline
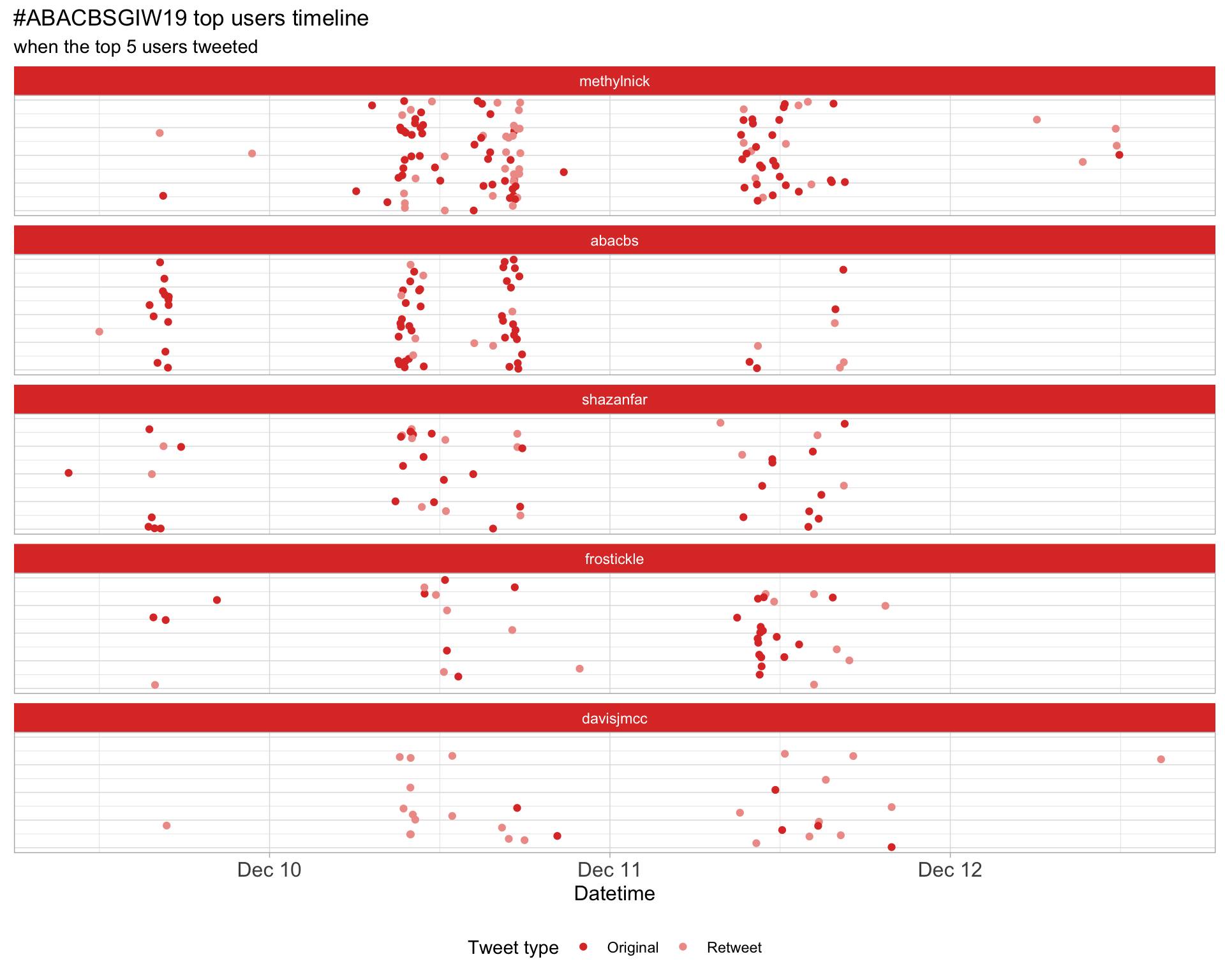
2.4 Top tweeters by day
Overall
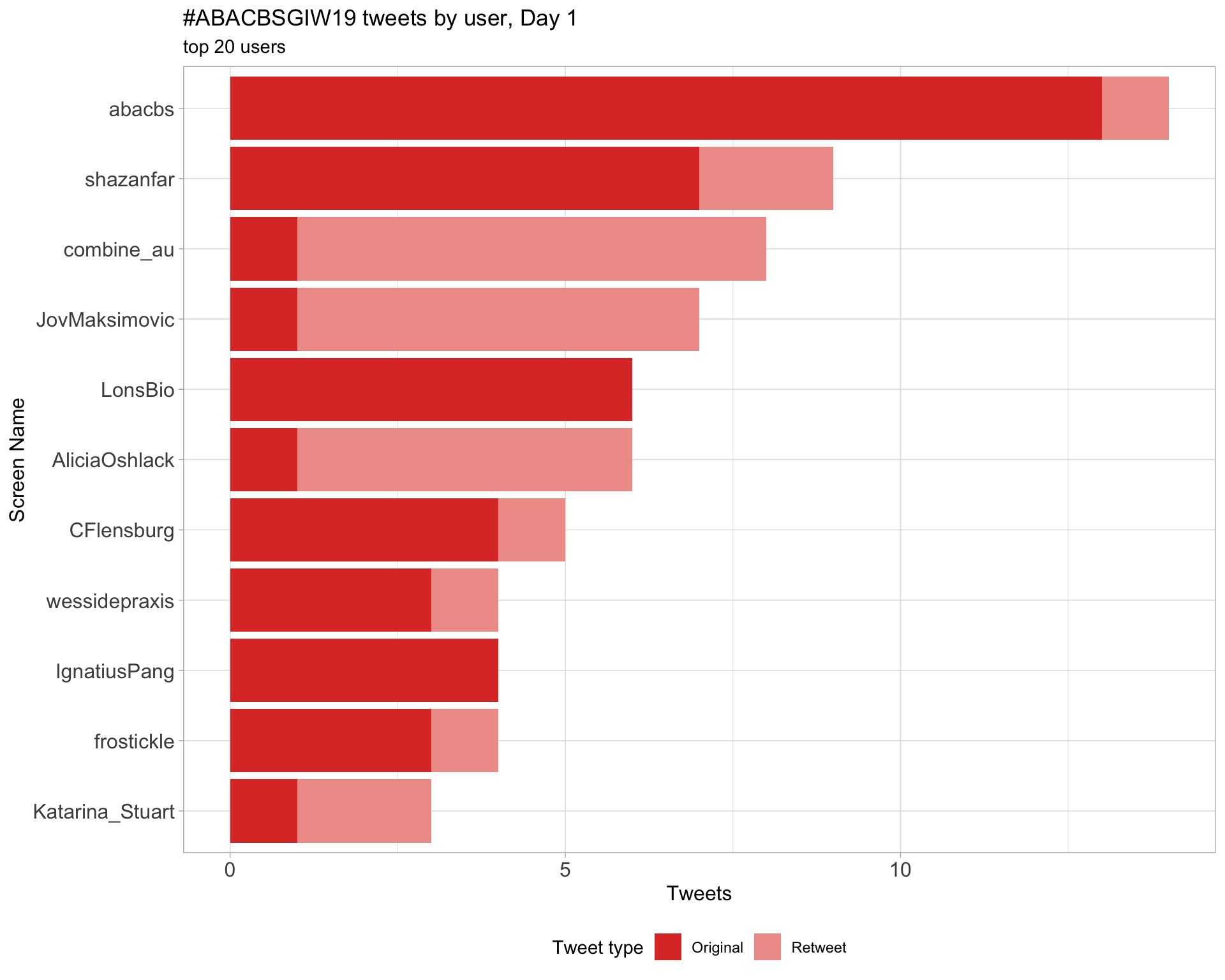
Day 1
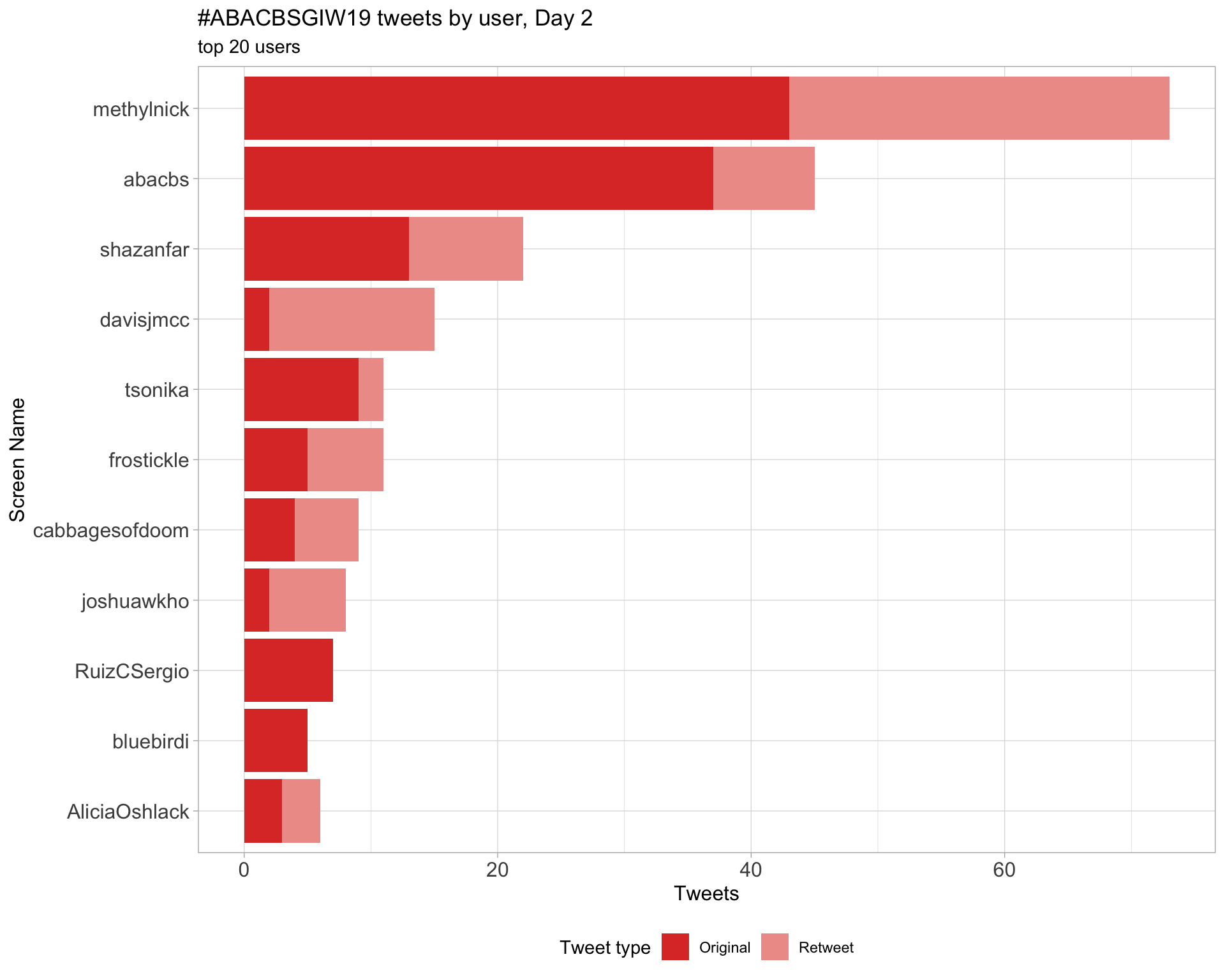
Day 2
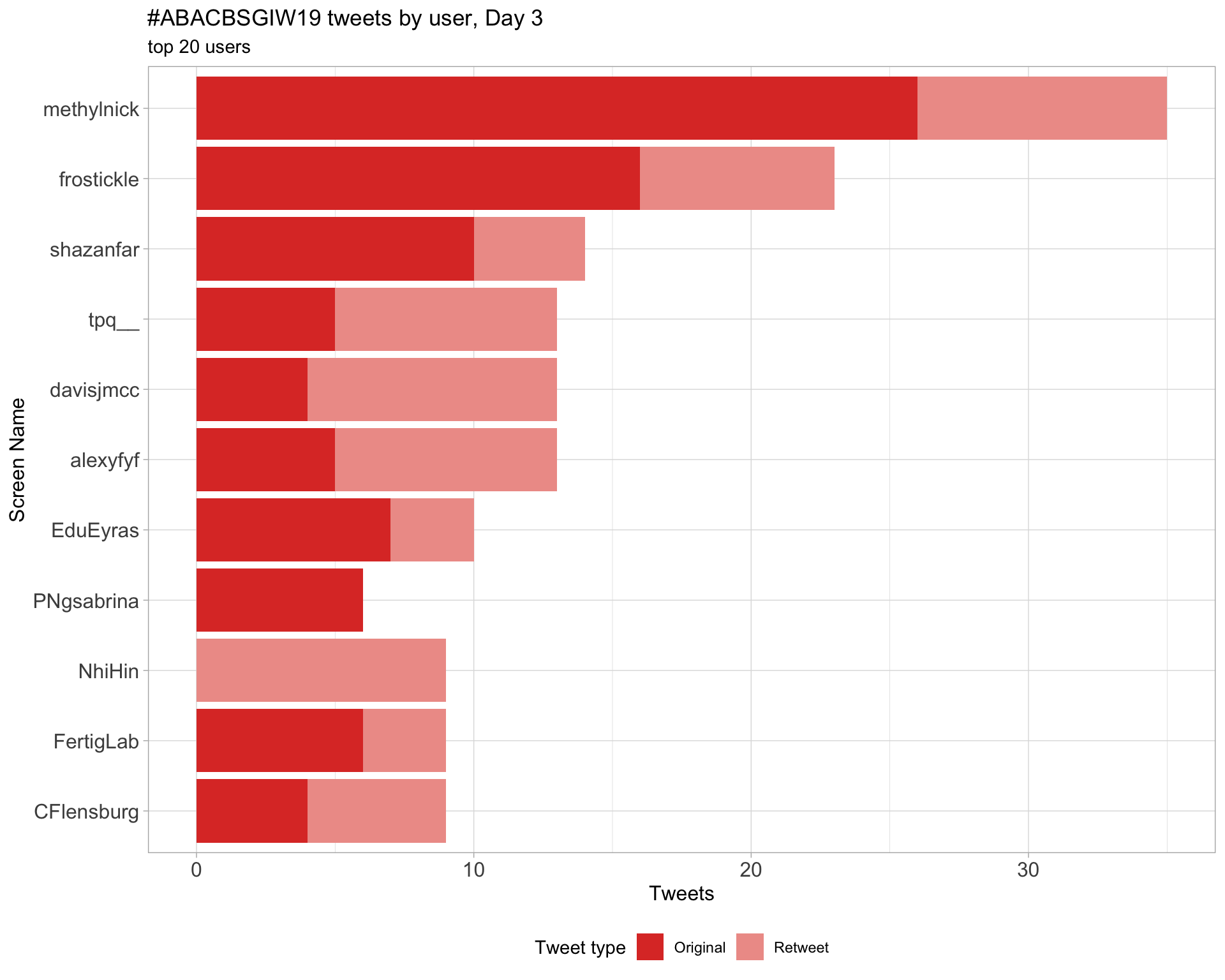
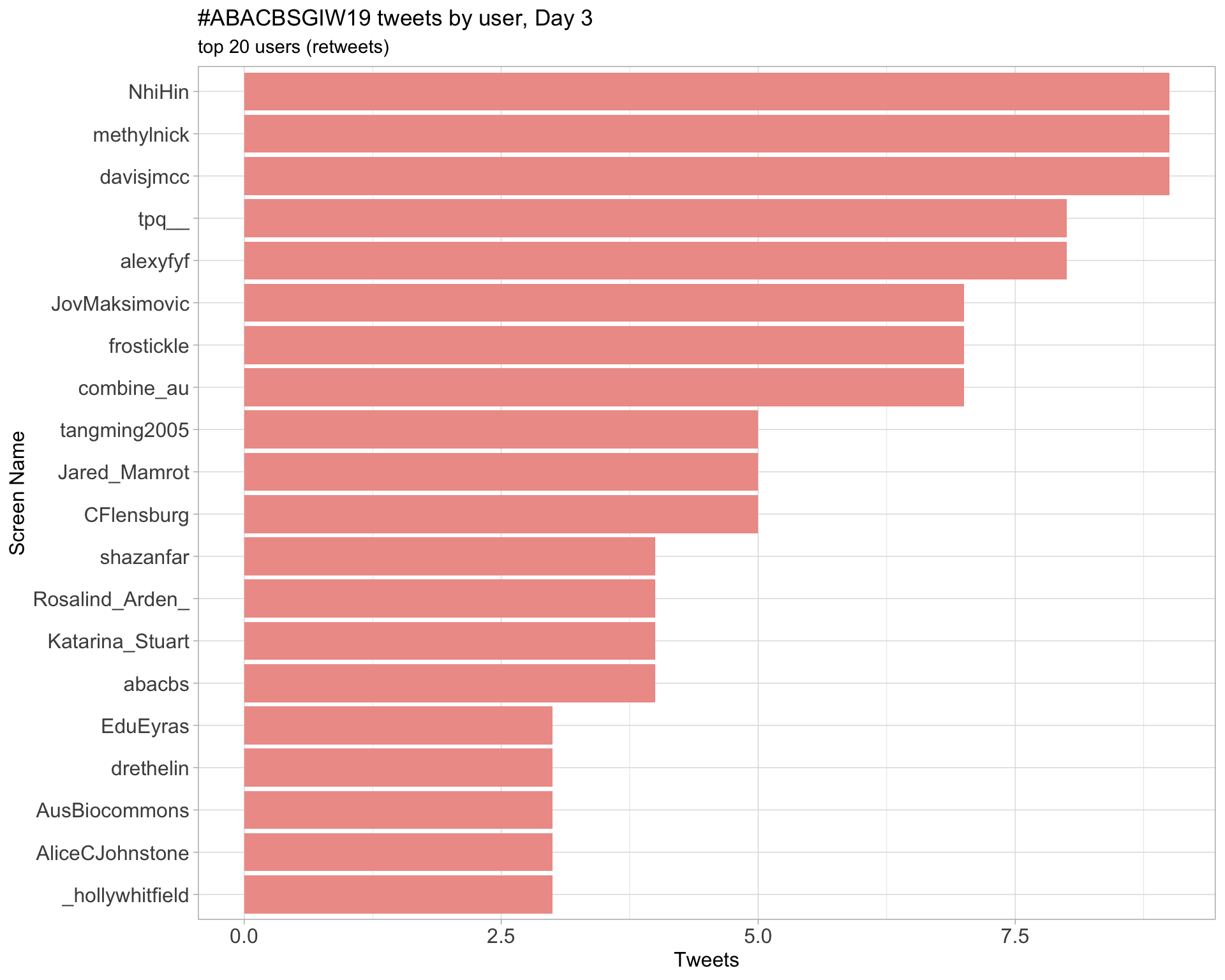
Day 3
Original
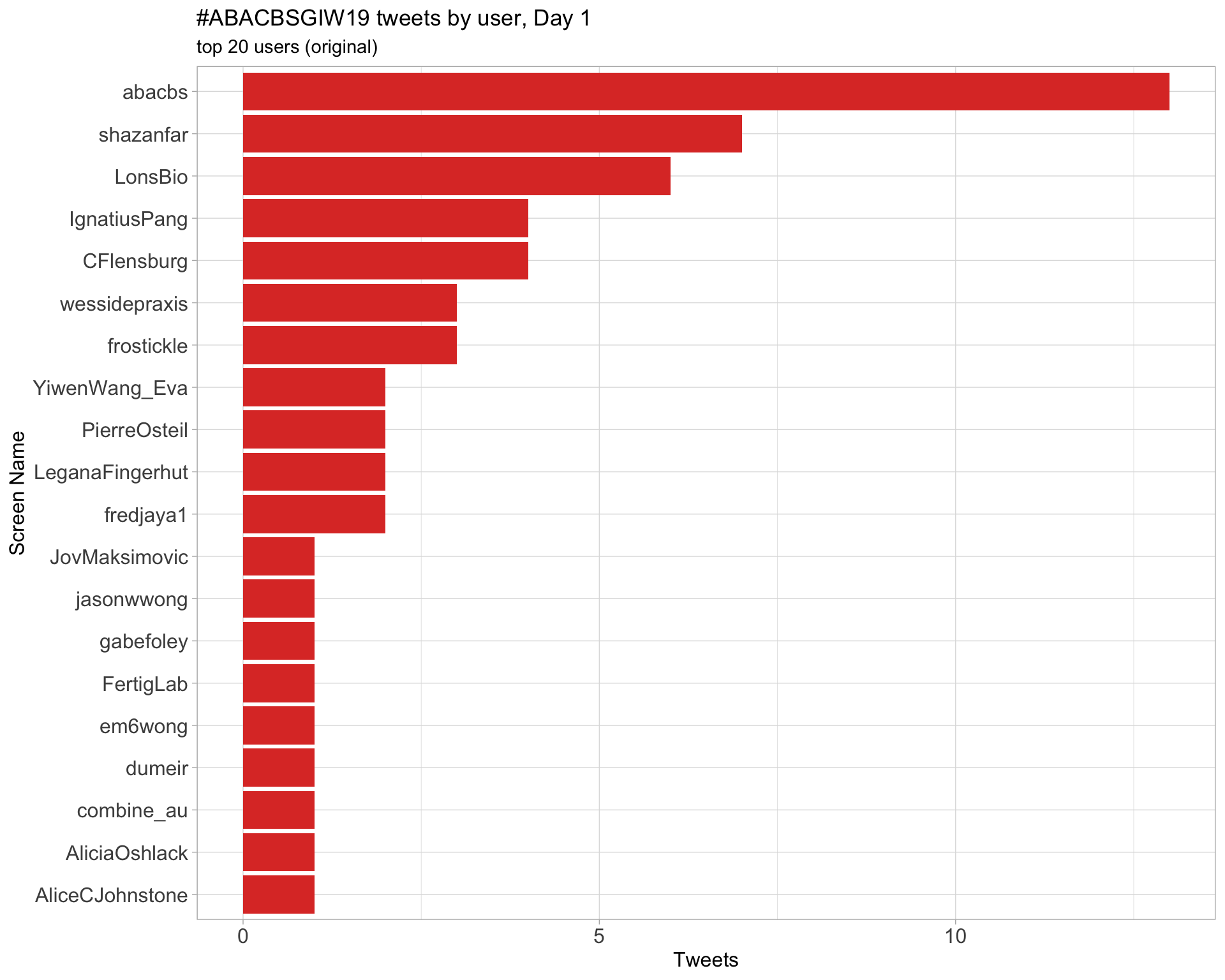
Day 1
Day 2
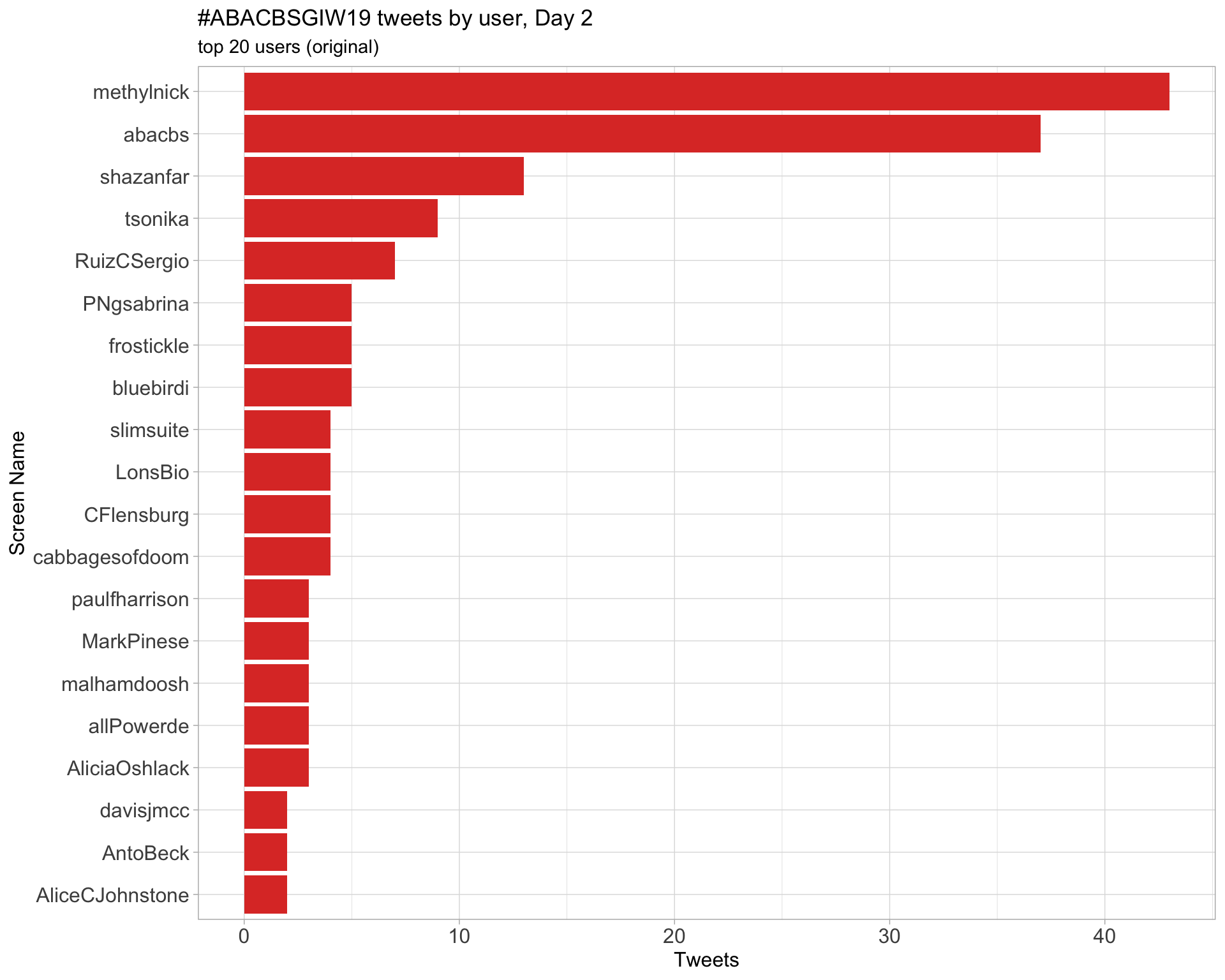
Day 3
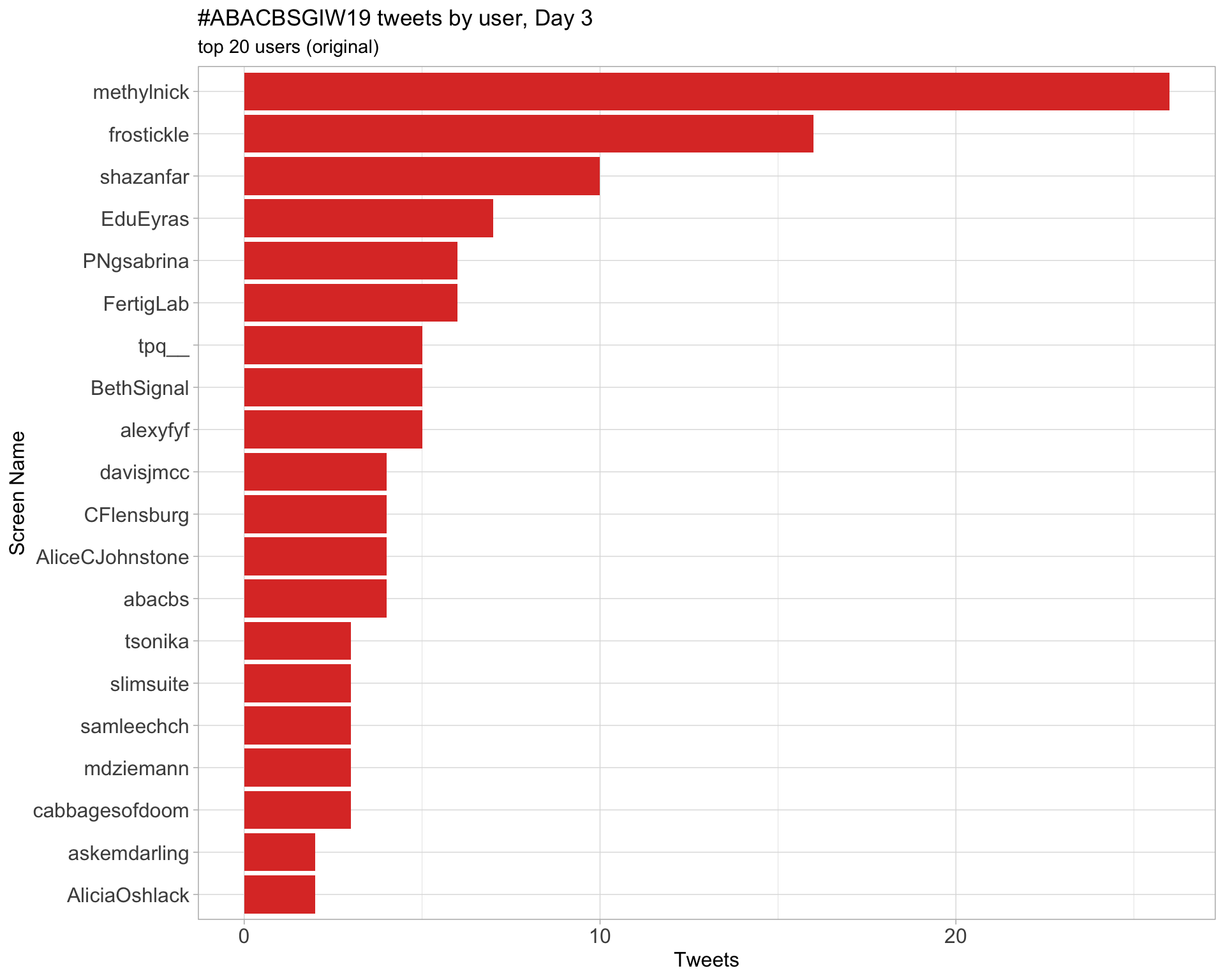
Retweets
Day 1
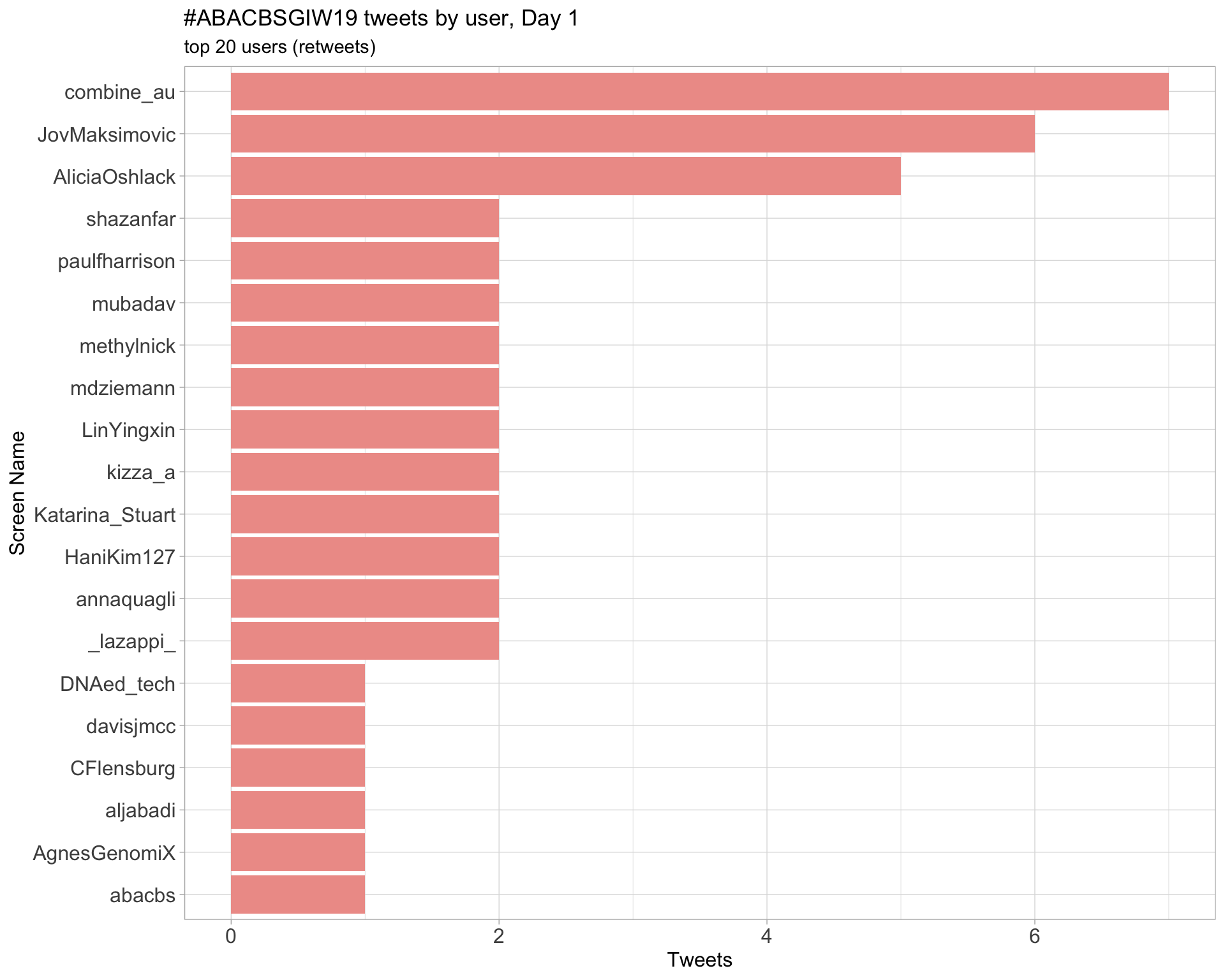
Day 2
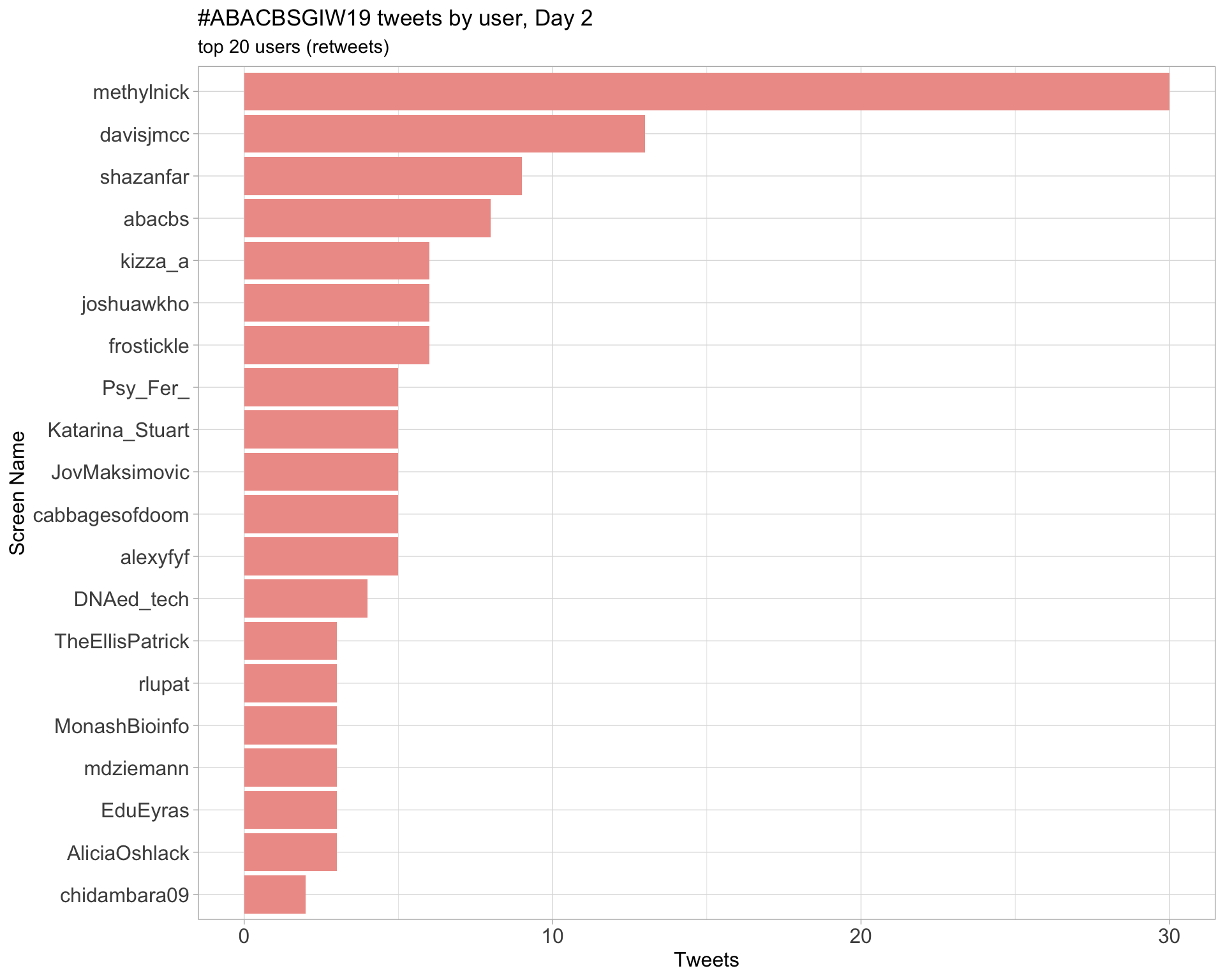
Day 3
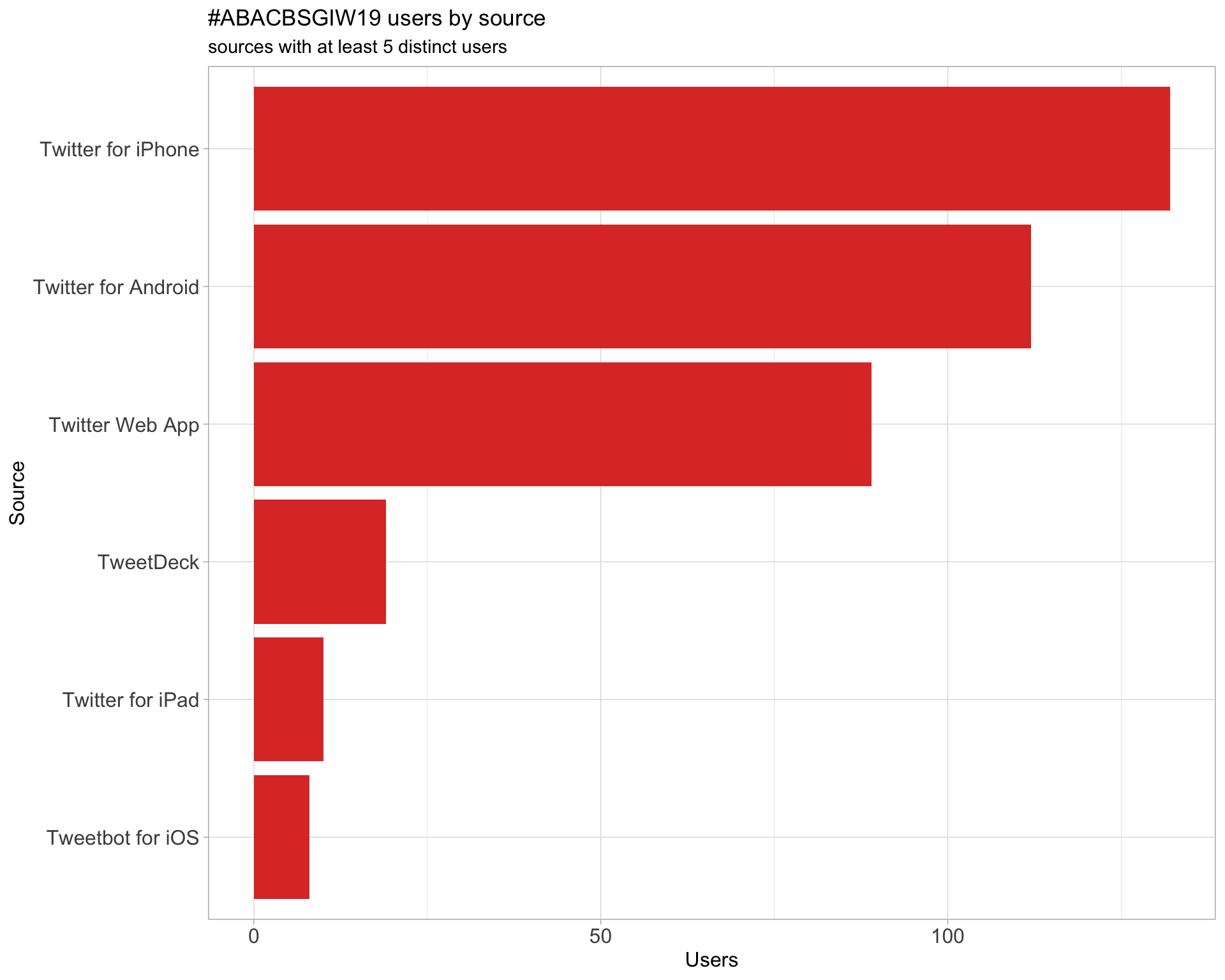
3 Sources
Users
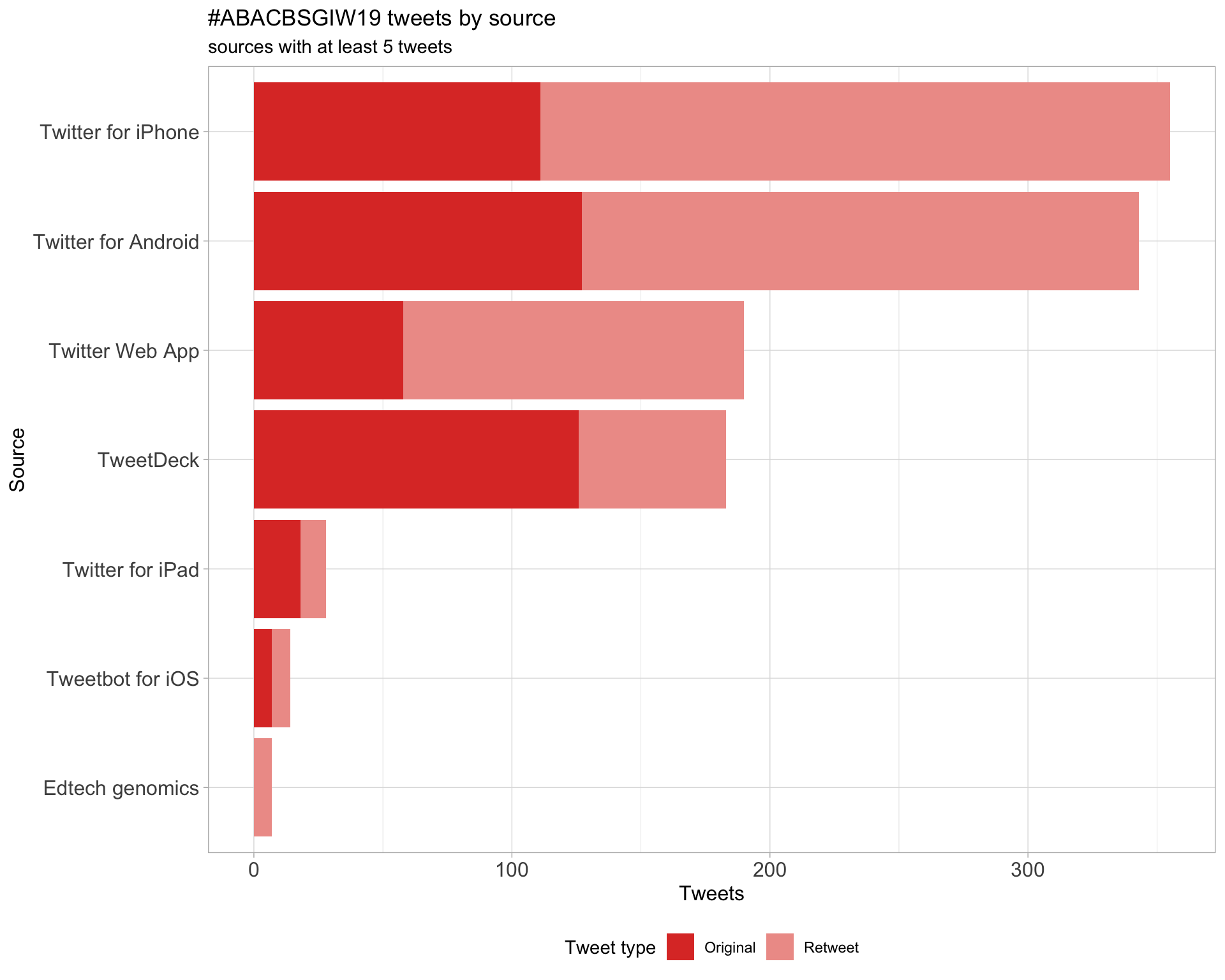
Tweets
4 Networks
4.1 Replies
The “replies network”, composed from users who reply directly to one another, coloured by PageRank.
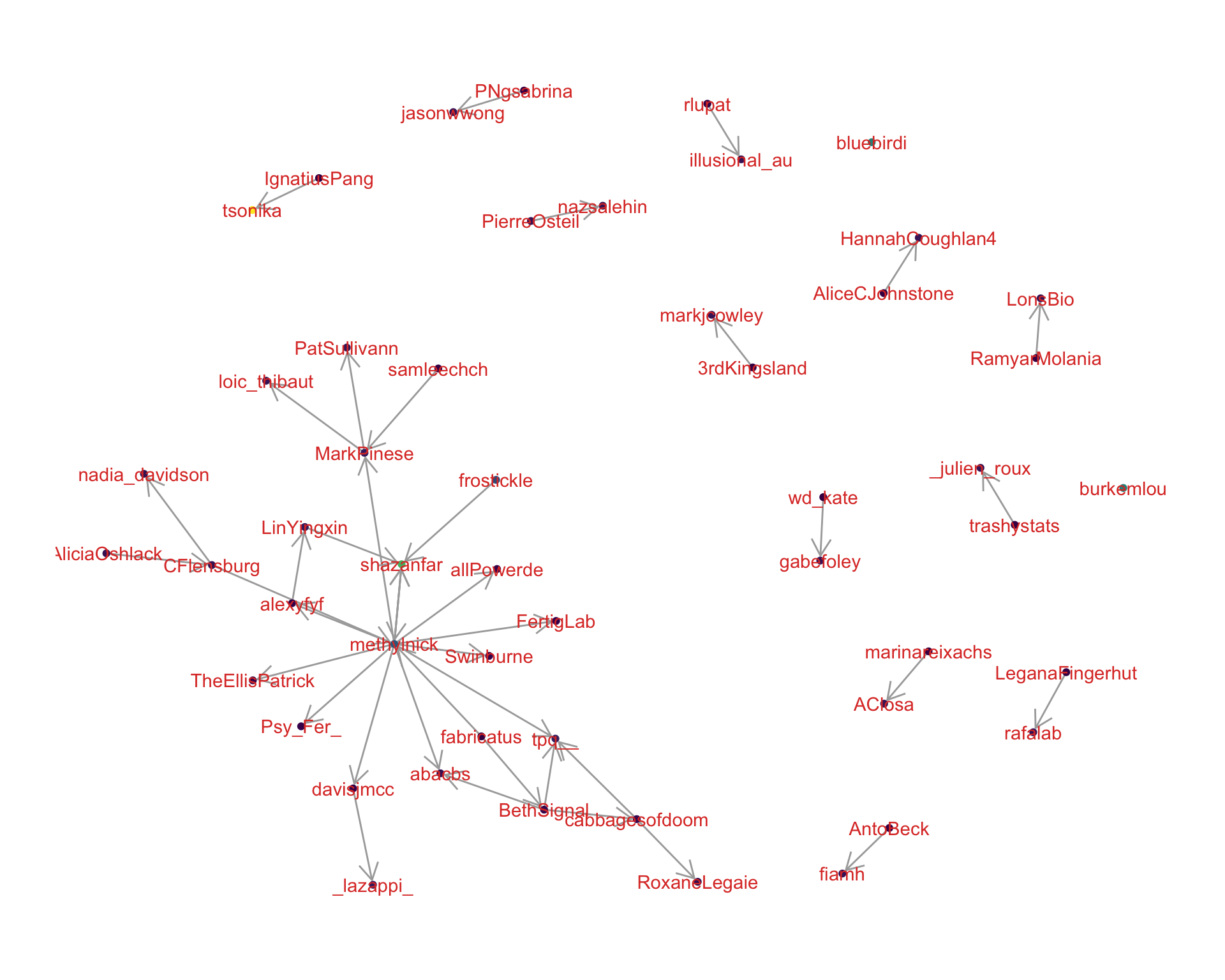
4.2 Mentions
The “mentions network”, where users mention other users in their tweets. Filtered for a k-core of 2. Node colour and size adjusted according to PageRank score.

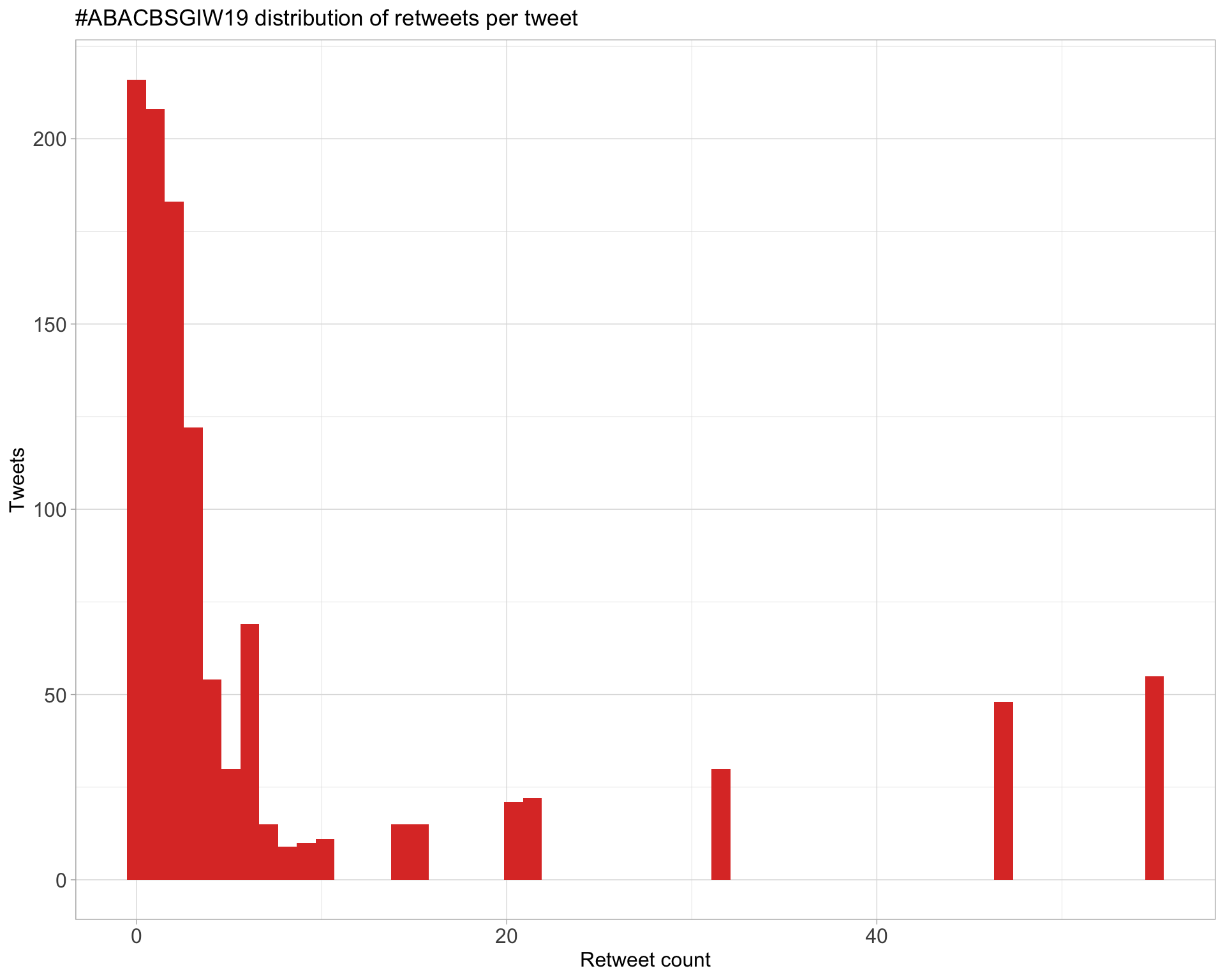
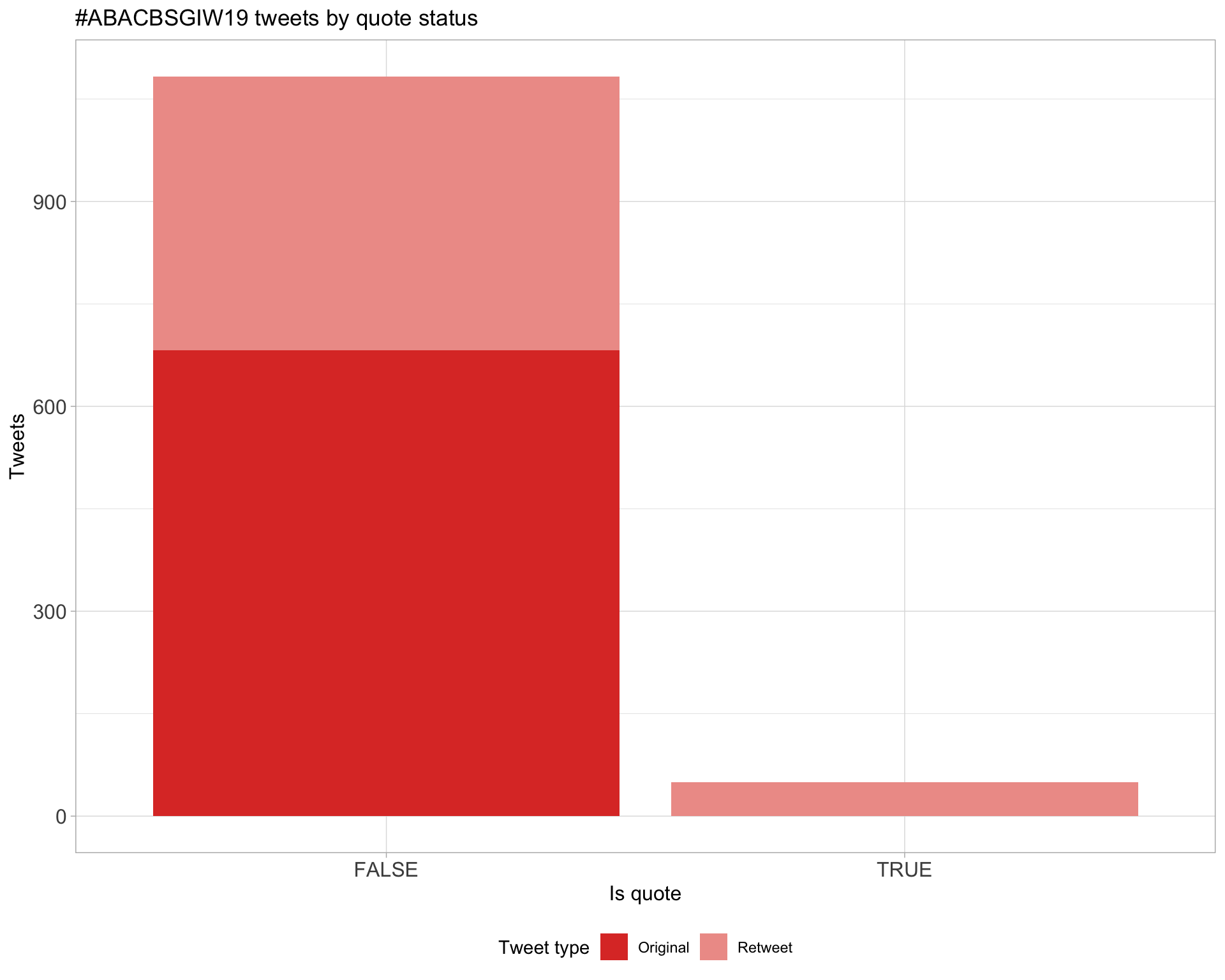
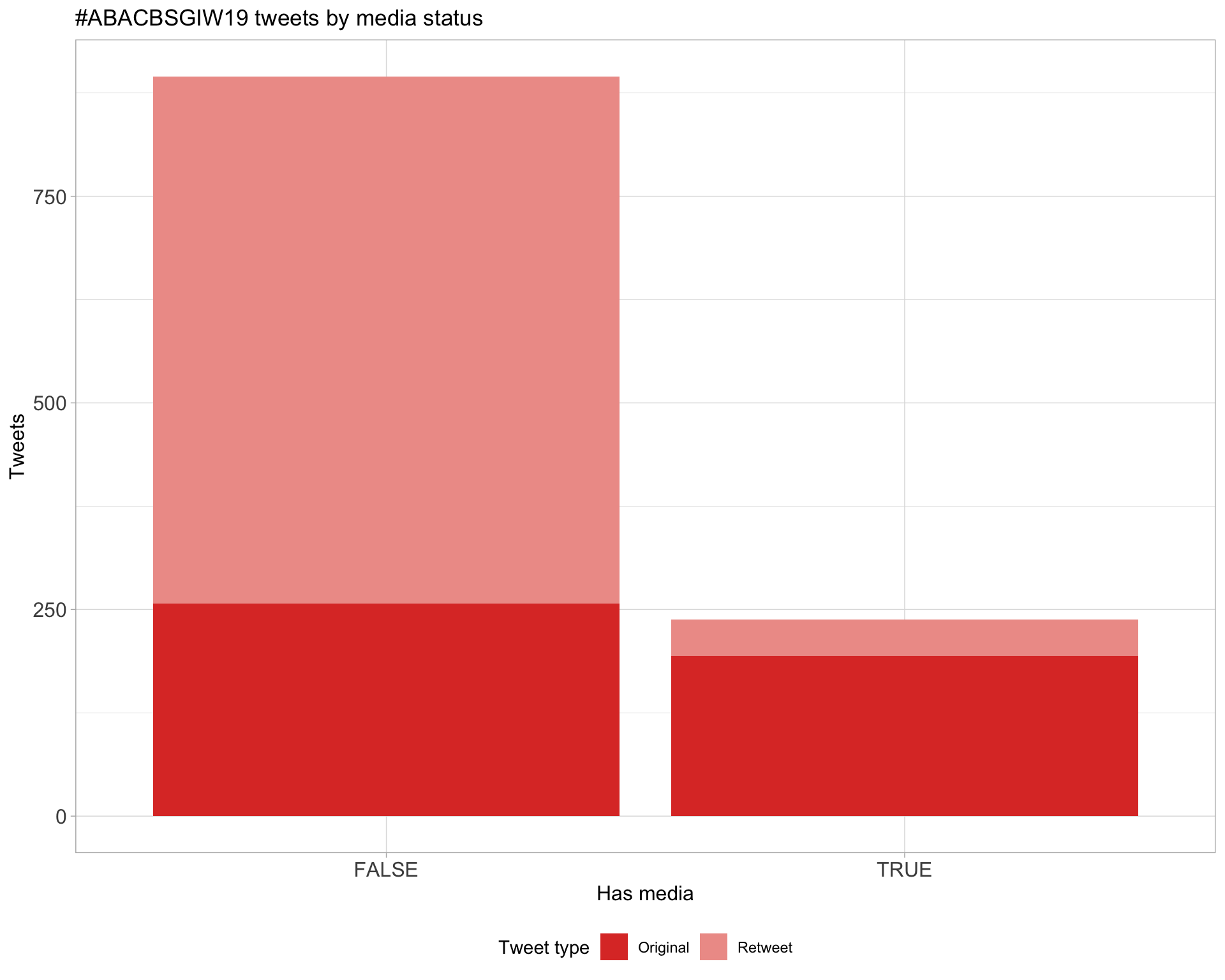
5 Tweet types
5.1 Retweets
Proportion
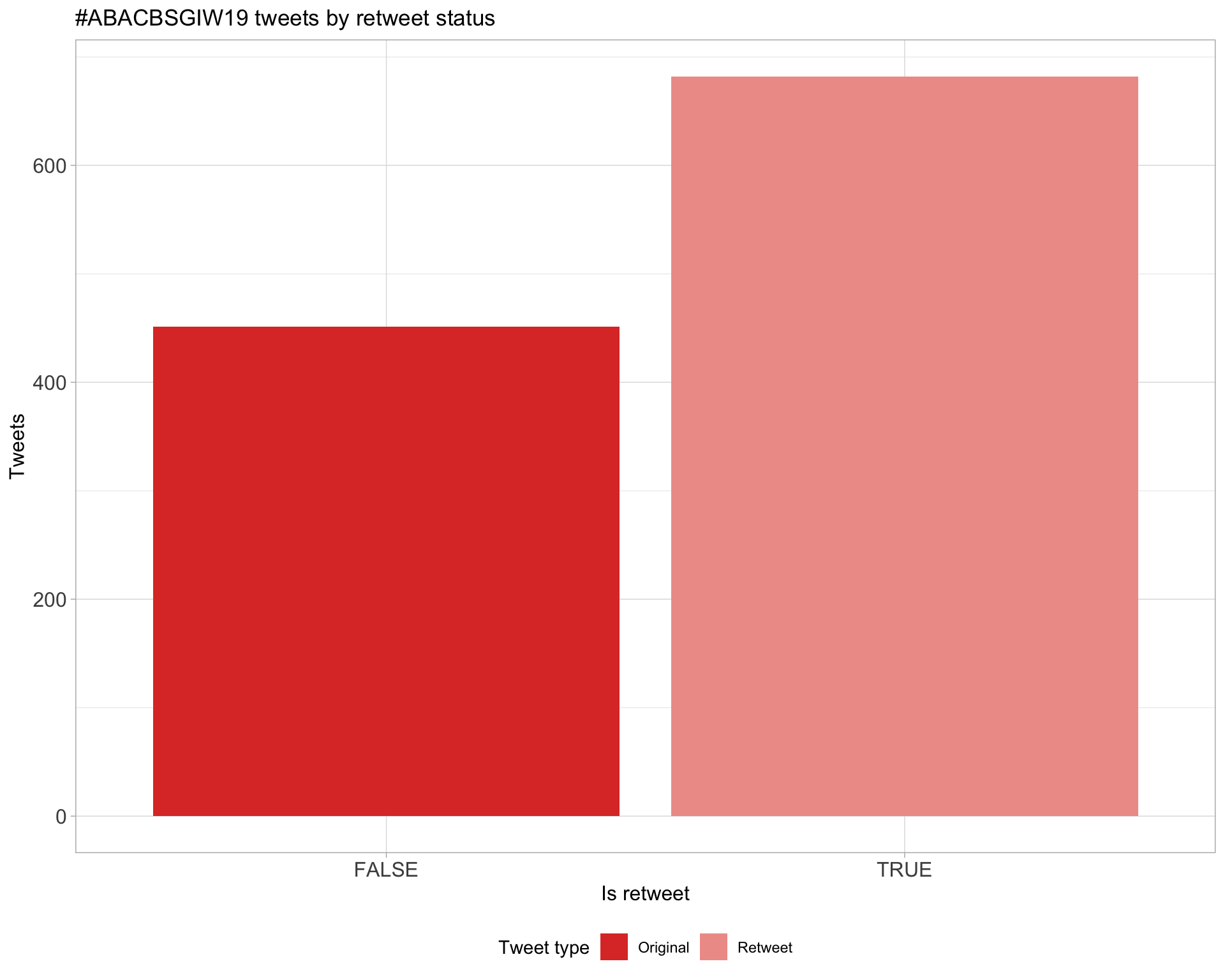
Count
Top 10
| screen_name | text | retweet_count |
|---|---|---|
| tpq__ |
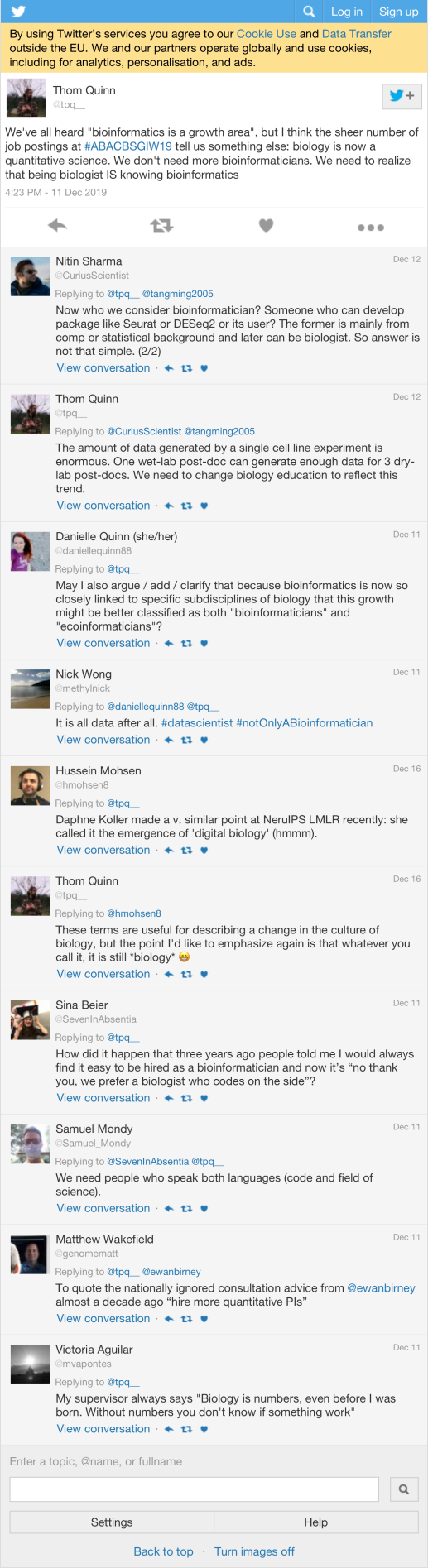
We’ve all heard “bioinformatics is a growth area”, but I think the sheer number of job postings at #ABACBSGIW19 tell us something else: biology is now a quantitative science. We don’t need more bioinformaticians. We need to realize that being biologist IS knowing bioinformatics |
55 |
| markjcowley | Hi #ABACBSGIW19 attendees, I’m hiring multiple #bioinformatics / #computationalbiology postdocs early next year in the areas of paediatric precision cancer medicine, large-scale genomic analysis on the cloud, noncoding variation, liquid biopsy, machine learning. DM me! | 47 |
| dgmacarthur | Hey #ABACBSGIW19 - interested in a postdoc, comp bio, stat gen, or software engineer position doing cool stuff with large-scale human sequencing data in Sydney or Melbourne, starting after May 2020? Drop me a line. Job descriptions going live early 2020! https://t.co/suFvy8KfRw | 32 |
| DrSimonHo |
We seek a Research Assistant with a background in bioinformatics or computational biology to join our lab at @Sydney_Uni This is a fixed-term (1 year) position and will be advertised in January. For further info and contact details visit https://t.co/gshu1EBC5P #ABACBSGIW19 https://t.co/jEPLn2ty4J |
21 |
| FertigLab | Shameless plug for anyone looking for a postdoc in single cell genomics and cancer immunotherapy at @hopkinskimmel @bloombergkimmel come find me at #ABACBSGIW19 https://t.co/J8RpLvawS1 | 20 |
| tpq__ | I’ve been thinking a lot lately about the social implications of AI research. Today, I’ll present our work on interpretable neural networks – “putting a window on the black box”. Beyond all the math and biology, this final slide summarizes my main purpose #ABACBSGIW19 https://t.co/TmNfLmjQfY | 15 |
| KelWyres | Enjoying #ABACBSGIW19 and want to pursue more exciting computational biology? We’re looking for a PhD student and a postdoc to work on new projects combining bacterial genomics and metabolic modeling. Check it out! PhD:https://t.co/4ydCJSmduD PostDoc:https://t.co/6cr8BCpBXa | 14 |
| MarkPinese | Guys, I don’t mean to be snarky but when we’re doing tool comparisons, can we please establish a ground truth first? Just because two tools agree with each other doesn’t mean they’re both right. Bias is by its nature consistent #ABACBSGIW19 | 10 |
| shazanfar | Elana Fertig @FertigLab opening Day 3 of #ABACBSGIW19 discussing factorisation techniques for high dimensional data such as single cell RNA-Seq. ProjectR ( https://t.co/honHMzcKDi ) performs transfer learning to extract interpretable gene features from identified NMF factors | 9 |
| rlupat | @illusional_au presenting Janis: a Python framework for portable pipelines at #ABACBSGIW19. Collaborations between @PeterMacRes @MelBioInf and @WEHI_research. https://t.co/cTSaBLCZat | 8 |
Most retweeted
5.2 Likes
Proportion
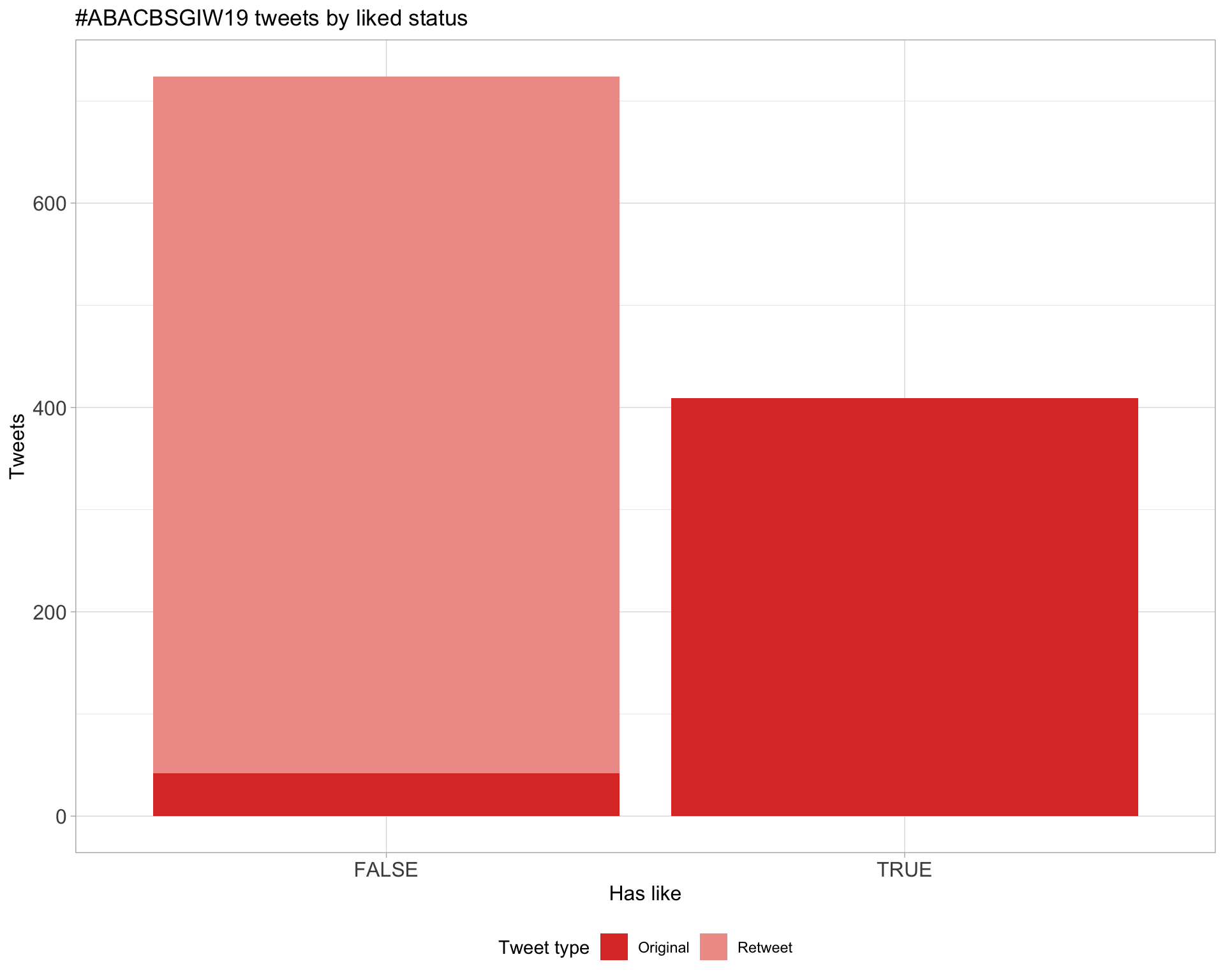
Count
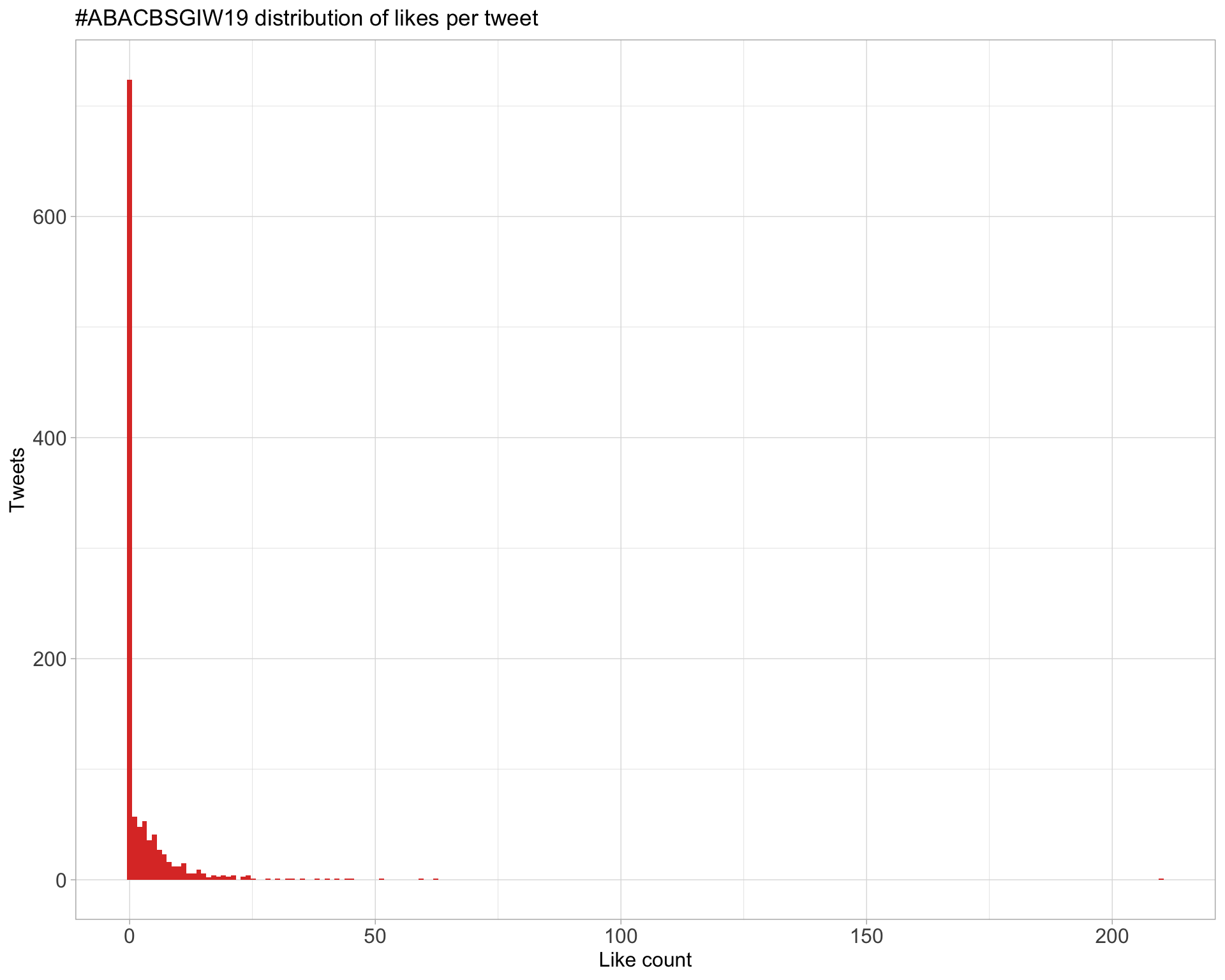
Top 10
| screen_name | text | favorite_count |
|---|---|---|
| tpq__ |
We’ve all heard “bioinformatics is a growth area”, but I think the sheer number of job postings at #ABACBSGIW19 tell us something else: biology is now a quantitative science. We don’t need more bioinformaticians. We need to realize that being biologist IS knowing bioinformatics |
210 |
| tpq__ |
Very honored to receive the “people’s choice award” for best talk at #ABACBSGIW19 – but must share the credit with my amazing team @adhambeyki @samleechch @truyenoz @DeakinA2I2 that made this work possible Check out our pre-print for the details 😁 https://t.co/A3bFEGsFzH |
62 |
| MarkPinese | Guys, I don’t mean to be snarky but when we’re doing tool comparisons, can we please establish a ground truth first? Just because two tools agree with each other doesn’t mean they’re both right. Bias is by its nature consistent #ABACBSGIW19 | 59 |
| AliciaOshlack | Congratulations @nadia_davidson for the @abacbs ECR award. This is well deserved recognition for all the awesome work you have done (check out all her work!) I know you’ll have an amazing career ahead as well #ABACBSGIW19 https://t.co/zG3Da8hMLu | 51 |
| markjcowley | Hi #ABACBSGIW19 attendees, I’m hiring multiple #bioinformatics / #computationalbiology postdocs early next year in the areas of paediatric precision cancer medicine, large-scale genomic analysis on the cloud, noncoding variation, liquid biopsy, machine learning. DM me! | 45 |
| RoxaneLegaie | Great turn out at the Professional Bioinformaticians get-together at #ABACBSGIW19 @abacbs 🤩 So… the question is… Who will be our Representative next year? 👇👤🕵️♀️ https://t.co/1B99PmM1hJ | 44 |
| davisjmcc |
#newpi reality check: I just got flagged down by a postdoc at the #ABACBSGIW19 conference dinner because……… He thought I was a waiter ……. |
42 |
| tpq__ | I’ve been thinking a lot lately about the social implications of AI research. Today, I’ll present our work on interpretable neural networks – “putting a window on the black box”. Beyond all the math and biology, this final slide summarizes my main purpose #ABACBSGIW19 https://t.co/TmNfLmjQfY | 40 |
| dgmacarthur | Hey #ABACBSGIW19 - interested in a postdoc, comp bio, stat gen, or software engineer position doing cool stuff with large-scale human sequencing data in Sydney or Melbourne, starting after May 2020? Drop me a line. Job descriptions going live early 2020! https://t.co/suFvy8KfRw | 38 |
| SHollizeck | If you didn’t get a chance to talk to me at my poster today, see me on Wednesday at number 150. #ABACBSGIW19 https://t.co/A2RpCIOPKh | 35 |
Most likes
5.3 Quotes
Proportion
Count
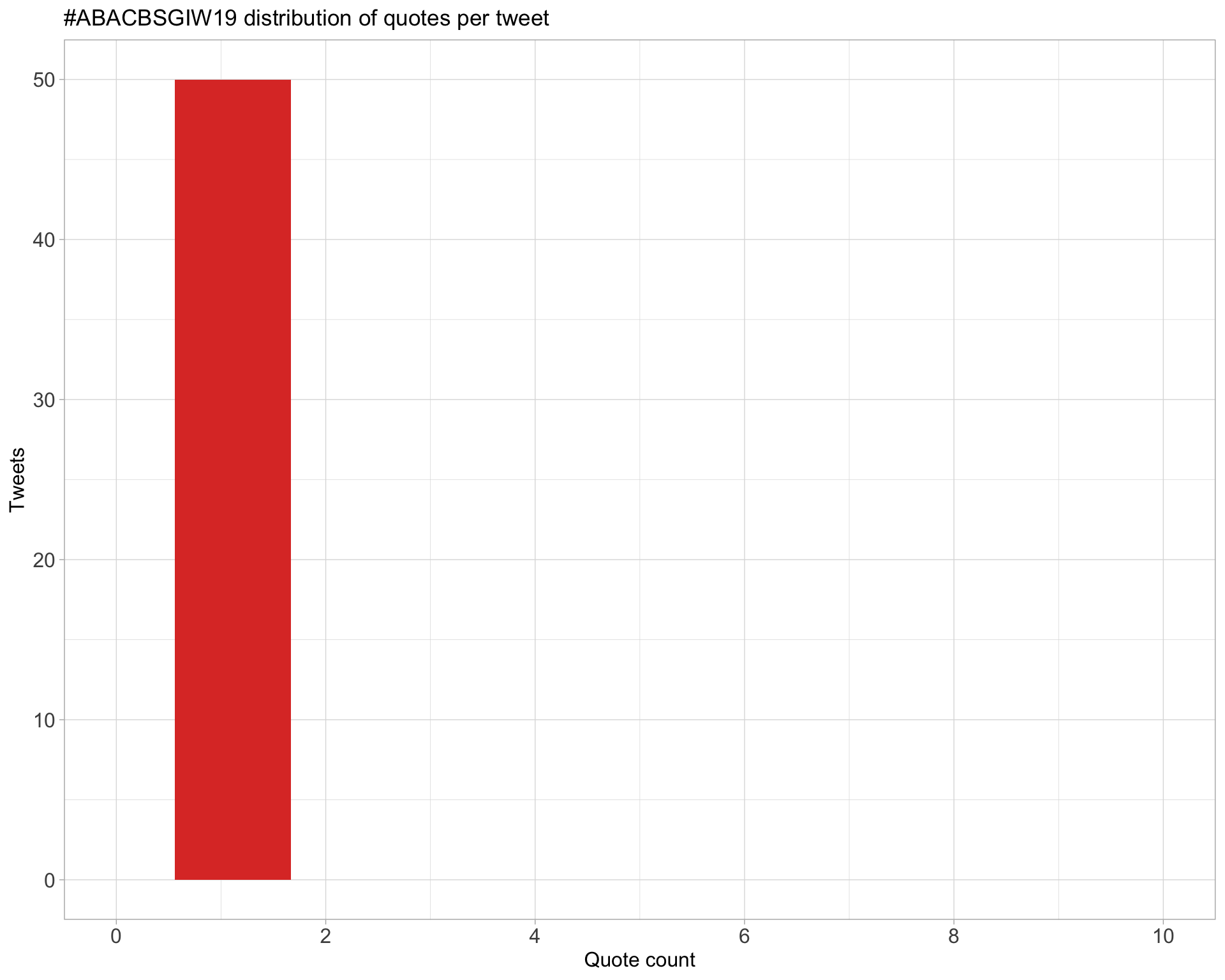
Top 10
| screen_name | text | quote_count |
|---|---|---|
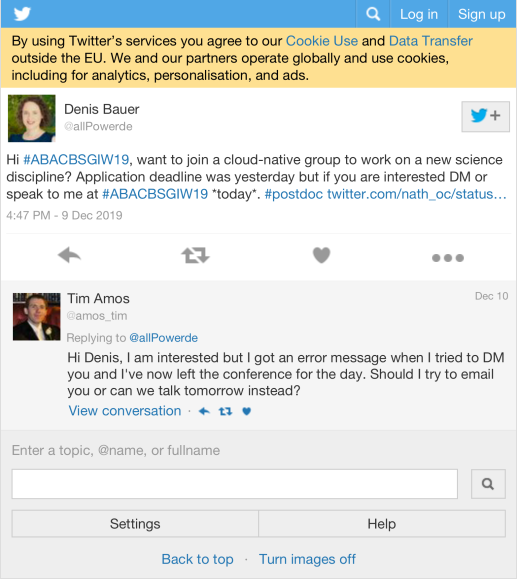
| allPowerde | Hi #ABACBSGIW19, want to join a cloud-native group to work on a new science discipline? Application deadline was yesterday but if you are interested DM or speak to me at #ABACBSGIW19 today. #postdoc https://t.co/YHfKo3ZKap | 1 |
| dgmacarthur | Hey #ABACBSGIW19 - interested in a postdoc, comp bio, stat gen, or software engineer position doing cool stuff with large-scale human sequencing data in Sydney or Melbourne, starting after May 2020? Drop me a line. Job descriptions going live early 2020! https://t.co/suFvy8KfRw | 1 |
| JovMaksimovic | The @abacbs postdoc survey is closing tomorrow at the end of the #ABACBSGIW19 conference. If you are a newbie postdoc (#combine19), a veteran postdoc or postdoc-identifying please complete the survey and let ABACBS know how they can best support and advocate for you. https://t.co/sIObXAFeEP | 1 |
| Al__Forrest | I think I missed this one #ABACBSGIW19 Reminder heaps of speaking slots available to be selected from the submitted abstracts! #ABACBSGIW19 is looking great BTW. https://t.co/Obi3Fq8k22 | 1 |
| LeganaFingerhut | My poster for #ABACBSGIW19 moved to spot 31! Come have a look! I’m happy to answer any questions you may have! @abacbs https://t.co/8bE1JKNfkd https://t.co/CL8cWXU6el | 1 |
| combine_au | Thanks @shazanfar! Yes we still have an amazing lineup of career panel! Please make your way to CPC main building level 6 after the opening session of #ABACBSGIW19 https://t.co/MXgOIZwQ8a | 1 |
| shazanfar | Good morning #ABACBSGIW19 ! Remember we are at Wallace Lecture theatre (map below) for the single stream part of the conference @abacbs https://t.co/KKkitTZ6Ev | 1 |
| shazanfar | How much is this a problem due to using transcriptomic readout as a proxy for surface protein abundance, in addition to general sparsity of Tx data? #ABACBSGIW19 @rafalab https://t.co/6V3Q4kkWVs | 1 |
| TheDawsonLab | For those of you who didn’t catch @SHollizeck talk during #VCBS2019 or at #COMBINE19. Come and see the awesome work he has done at poster 150 during the lunch break! #ABACBSGIW19. https://t.co/5xA00C1XAb | 1 |
| tsonika | Tyrone and Javier would love to discuss their work with you. Go say hello! #ABACBSGIW19 #COMBINE19 https://t.co/f22R3xhWcK | 1 |
Most quoted
6 Media
Proportion
Top 10
| screen_name | text | favorite_count |
|---|---|---|
| AliciaOshlack | Congratulations @nadia_davidson for the @abacbs ECR award. This is well deserved recognition for all the awesome work you have done (check out all her work!) I know you’ll have an amazing career ahead as well #ABACBSGIW19 https://t.co/zG3Da8hMLu | 51 |
| RoxaneLegaie | Great turn out at the Professional Bioinformaticians get-together at #ABACBSGIW19 @abacbs 🤩 So… the question is… Who will be our Representative next year? 👇👤🕵️♀️ https://t.co/1B99PmM1hJ | 44 |
| tpq__ | I’ve been thinking a lot lately about the social implications of AI research. Today, I’ll present our work on interpretable neural networks – “putting a window on the black box”. Beyond all the math and biology, this final slide summarizes my main purpose #ABACBSGIW19 https://t.co/TmNfLmjQfY | 40 |
| SHollizeck | If you didn’t get a chance to talk to me at my poster today, see me on Wednesday at number 150. #ABACBSGIW19 https://t.co/A2RpCIOPKh | 35 |
| lnly0311 | Thanks @combine_au for giving me the best poster presentation😆! Today’s career panel and social night are definitely excellent. Big shout out to the symposium organisers for such a good day. Looking forward to the next two days at #ABACBSGIW19 conference! https://t.co/eolNglQINO | 33 |
| YiwenWang_Eva | Thanks for the chance to present my work #combine19 My poster is packed up and ready for ABACBS. 😉#ABACBSGIW19 https://t.co/i3l4anaUXH | 32 |
| bluebirdi | As I tweet from the air, a final thanks to all the amazing people I met in Sydney over the last week and a half. Thanks so much for all the chats and support - from the undergrads to the seniors! See you at the next bioinformatics gathering!! #bioinfosummer #ABACBSGIW19 https://t.co/ZtVwtlCjmo | 28 |
| wessidepraxis | An action shot of our amazing @UofABioinfoHub PhD student @NhiHin doing her @combine_au winning talk for the full @abacbs audience #ABACBSGIW19 https://t.co/Q9lKMZGAzT | 25 |
| methylnick | This river plot is next level. @MarioniLab @abacbs #ABACBSGIW19 #visualisation @MonashBioinfo https://t.co/SuAZL98LaE | 24 |
| methylnick | Lightning talk by @Psy_Fer_ showcasing a swag of tools he has created for @nanopore data. A lot for 90 secs. @abacbs #ABACBSGIW19 https://t.co/G1JCrfvmAh | 24 |
6.1 Most liked image

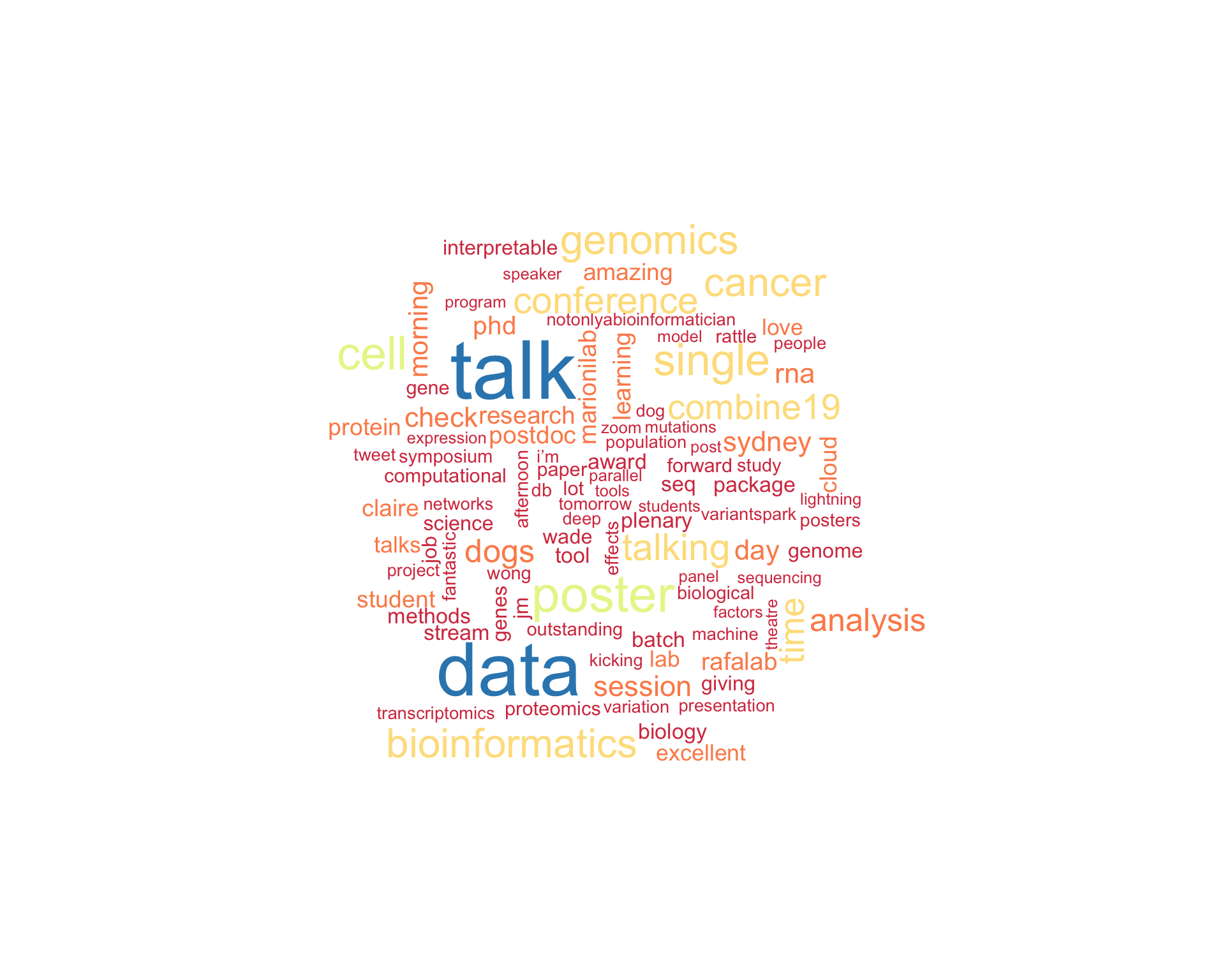
7 Tweet text
7.1 Word cloud
The top 100 words used 3 or more times.
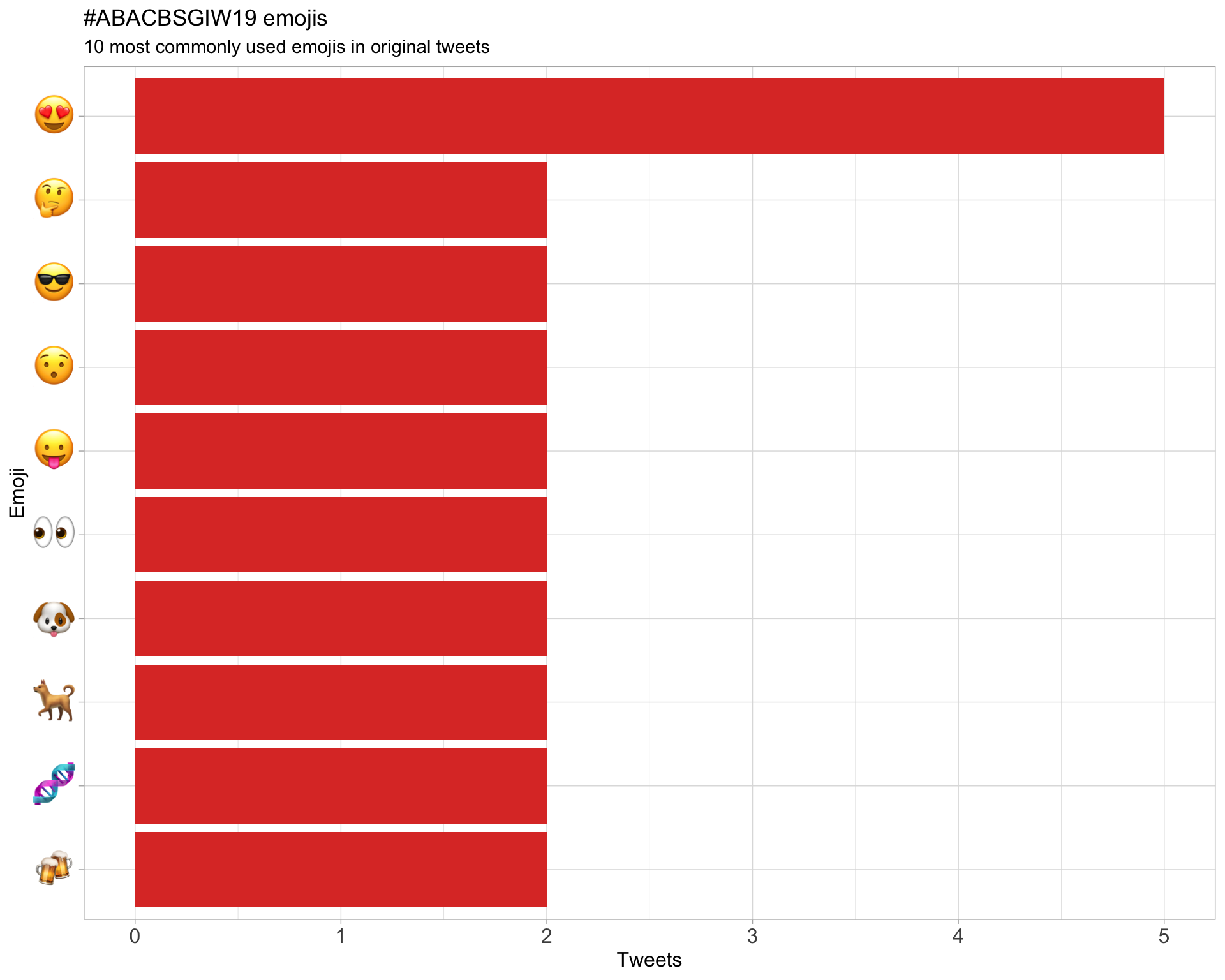
7.3 Emojis
7.4 Bigram graph
Words that were tweeted next to each other at least 3 times.
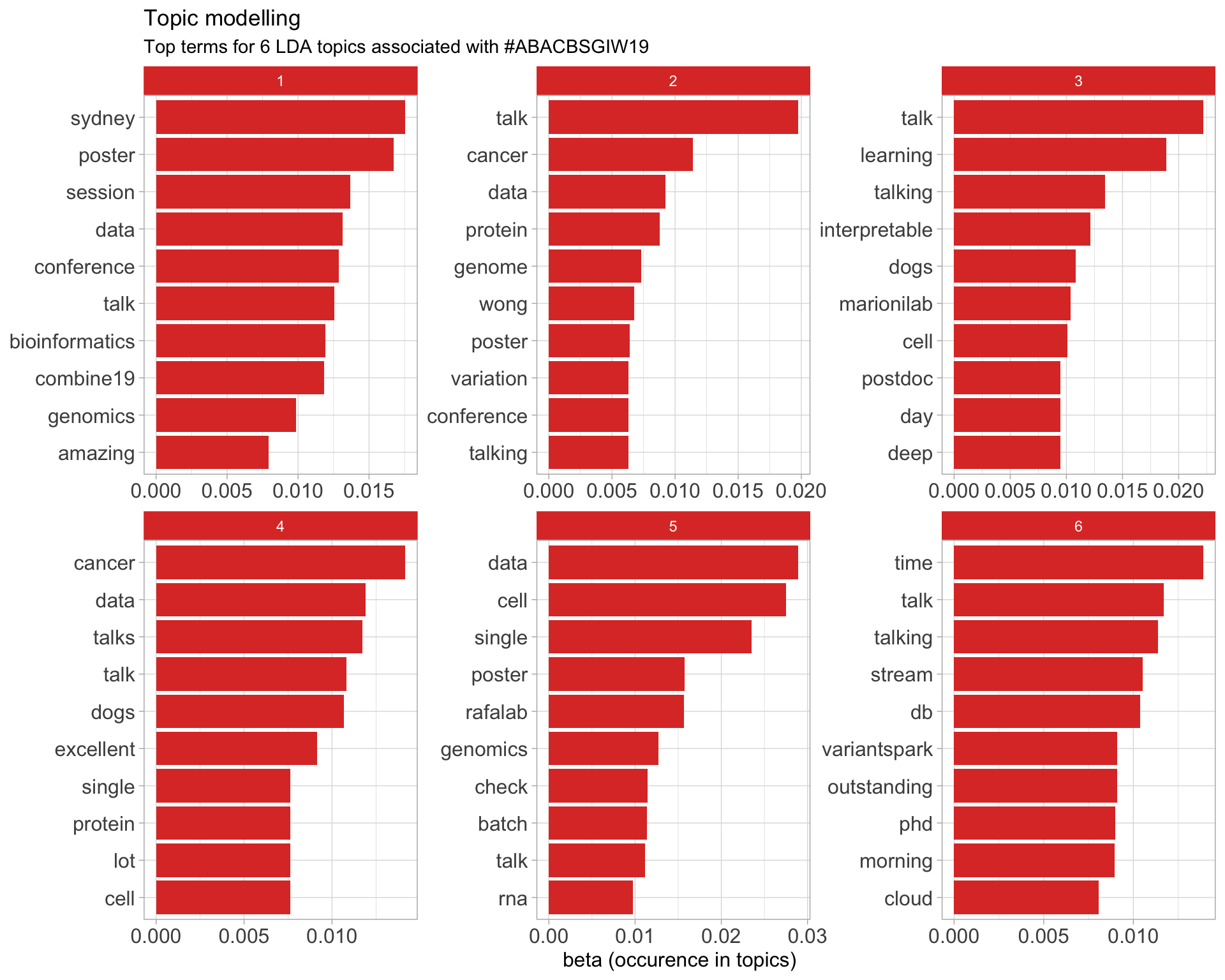
7.5 Topic modelling
Top 10 words associated with 6 topics identified by LDA.
7.5.1 Representative tweets
Most representative tweets for each topic
Topic 1
| screen_name | text | gamma |
|---|---|---|
| dgmacarthur | Hey #ABACBSGIW19 - interested in a postdoc, comp bio, stat gen, or software engineer position doing cool stuff with large-scale human sequencing data in Sydney or Melbourne, starting after May 2020? Drop me a line. Job descriptions going live early 2020! https://t.co/suFvy8KfRw | 0.9941808 |
| frostickle |
Michael Franklin from the @PeterMacCC/@PeterMacRes talking to us about Portable Python Pipelines. “Janis” is a Python framework with a simple API & registry of tools and types that generates CWL and WDL pipelines. #ABACBSGIW19 https://t.co/3b8uUrESe7 |
0.9936559 |
| methylnick | This is why I love what I do, the opensource community and presentation of top science at @abacbs #ABACBSGIW19 where I heard about Gencore. @biorxivpreprint paper + @github repo are here. https://t.co/mGufpGg7dQ https://t.co/15Mr6rAiUc install onto our server and run. #cleaner https://t.co/MGTHRwSi5o | 0.9926630 |
| methylnick | Love this #UMAP projection. It’s art. And science. Beginning to warm up to #singleCell experiments. If done well. EF using projects to go back to a reference to find new factors. In this case, gender as the samples were not sexed before running. @abacbs #ABACBSGIW19 https://t.co/ghJ4wpXNBG | 0.9922591 |
| methylnick | Great anaolgy by @allPowerde for the value proposition for #inNubium with car ownership and ride share. The #Uber for #genomics. @abacbs #ABACBSGIW19 less than a cup of coffee to serve data in cloud per day. #coffeeEconomy https://t.co/oFGbBkHLuj | 0.9918083 |
| bluebirdi | As I tweet from the air, a final thanks to all the amazing people I met in Sydney over the last week and a half. Thanks so much for all the chats and support - from the undergrads to the seniors! See you at the next bioinformatics gathering!! #bioinfosummer #ABACBSGIW19 https://t.co/ZtVwtlCjmo | 0.9918083 |
| bluebirdi | While I cant be at the @PMV_Australia symposium today @WEHI_research, I hope everyone has fantastic time! So many amazing talks - Remember it starts at 1pm! Anyone at #ABACBSGIW19 - feel free to sign up for the next one! https://t.co/9JXH3O79tf #proteomics #metabolomics | 0.9918083 |
| trombleyel | #h3h3isoverparty #MissUniverse2019 #LakeShow TekniKool Air Conditioning Sydney Installation & Service - Daikin Air Conditioning Sydney #ABACBSGIW19 #CrisisOnInfiniteEarths #RIPJUICEWRLD https://t.co/VZm3tFjl5E | 0.9913016 |
| minouye271 | At #ABACBSGIW19 and looking for a postdoc? Please do get in touch https://t.co/R5AQWOXxif (or talk to @RuizCSergio at the conf). We have openings at our Cambridge node in comp genomics (https://t.co/7z1fYm7a8z) & structural genomics (forthcoming), and at our Melb node in 2020. | 0.9913016 |
| watsonhaigh | #Sydney bound to give a half day #Snakemake workshop following the @abacbs national #bioinformatics conference in Australia. Let me know if you’re one of those attending the workshop and what level of exposure you’ve already had with Snakemake #ABACBSGIW19 @johanneskoester | 0.9913016 |
Topic 2
| screen_name | text | gamma |
|---|---|---|
| PaulLacaze | (seeing everyone else is) PhDs, RAs, post doc positions now recruiting w Monash @ the ASPREE STUDY, Alfred hospital #ABACBSGIW19. Bfx, epi, biostats, comp genetics, systems biol, data science. World-class genome-phenome dataset. DM me for details @Monash_SPHPM @ASPREE_aus | 0.9951783 |
| davisjmcc | One of my #ABACBSGIW19 highlights earlier today was casually sitting in the front grow of PuXue Qiao’s talk and nodding along furiously. Very proud of the outstanding job she did delivering a talk on behalf of Shifu Chen who couldn’t attend due to visa issues | 0.9939297 |
| tsonika | MOFA: “computational method for discovering the principal sources of variation in multi‐omics data sets. MOFA infers a set of (hidden) factors that capture biological and technical sources of variability” https://t.co/NUxggDvqPz #ABACBSGIW19 | 0.9936559 |
| davisjmcc | Xining Li gave a fantastic talk on A-to-I editing in RNA. Elegant experimental system and thoughtful bioinformatic analyses from his Masters(!) project with Alistair Chalk and Carl Walkley at @SVIResearch - he’s on the market for RA and PhD opportunities! #ABACBSGIW19 | 0.9936559 |
| shazanfar | Love the map on the left including important Christmas tree landmark 😊 #ABACBSGIW19 is a victim of its own success meaning we need to 🚶♂️🚶♀️ over to larger nearby Wallace lecture theatre (WT) for the single stream/plenary sessions https://t.co/z5wL3DXVyf | 0.9933562 |
| tpq__ | I’ve been thinking a lot lately about the social implications of AI research. Today, I’ll present our work on interpretable neural networks – “putting a window on the black box”. Beyond all the math and biology, this final slide summarizes my main purpose #ABACBSGIW19 https://t.co/TmNfLmjQfY | 0.9933562 |
| PNgsabrina | @jasonwwong on determinants of CTCF/cohesin binding site mutagenesis in cancer genomes. Interesting to learn the relationships between mutation and DNA methylation in colorectal cancer 😯 And a beautiful Circos plot 😍 #ABACBSGIW19 https://t.co/iL4ys7iW2o | 0.9926630 |
| dumeir | Started #COMBINE19 with an excellent presentation by my friend @rfroum and ended with an insightful career panel by @rafalab, @MarcelDinger, Dr Denis Bauer and Dr Emily Wong. Thank you @combine_au for allowing me to be an abstract reviewer and see you tomorrow for #ABACBSGIW19! | 0.9926630 |
| JasonJinxin | Many thanks for @abacbs Organizing committee. Such a great opportunity for me to talk my current genome-metabolic modelling research at #ABACBSGIW19 and nice discussion with Eva on integrating the time series omics data with GSMM. https://t.co/JbbUm7AopN | 0.9926630 |
| abacbs | JM: Multi-omics factor analysis (MOFA). Looking at variation which is shared between methylation, chromatin accessibility and RNA expression. Demethylation and accessibility of enhancers is defining lineages. #ABACBSGIW19 | 0.9922591 |
| FertigLab | Jason Wong presenting really interesting work on interactions between mutation and Epigenetics - The interaction between cytosine methylation and processes of DNA replication and repair shape the mutational landscape of cancer genomes. #ABACBSGIW19 https://t.co/o0KwhwyksQ | 0.9922591 |
Topic 3
| screen_name | text | gamma |
|---|---|---|
| shazanfar | Elana Fertig @FertigLab opening Day 3 of #ABACBSGIW19 discussing factorisation techniques for high dimensional data such as single cell RNA-Seq. ProjectR ( https://t.co/honHMzcKDi ) performs transfer learning to extract interpretable gene features from identified NMF factors | 0.9941808 |
| shazanfar |
Excellent vis to understand this huge dataset! @omnislip (recently graduated!) PhD student in @MarioniLab led this analysis & put a huge amount of work into ensuring the data is accessible and interpretable #ABACBSGIW19 Paper https://t.co/xaRaWbojP2 App https://t.co/mzGgnCuSeQ https://t.co/n0CnVv3Xpt |
0.9941808 |
| frostickle |
Claire mainly recruits recruits Kelpies from the general population. There are a few distinct types of working dog. “Yard dogs”, “3-sheep trial dogs” and “paddock dogs”. These dogs have an extraordinary skills in independent problem solving when performing maneuvers #ABACBSGIW19 https://t.co/HukVXuL9Jj |
0.9939297 |
| frostickle | “What makes dogs so good for studying is cuz they’ve been bred down a tight bottleneck, we removed a lot of the background genes. So within a breed you get a quieting of the noise, e.g. the major gene for height is so prominent you could visually identify it.” #ABACBSGIW19 https://t.co/wj7iHbFGTp | 0.9939297 |
| frostickle |
Great talk by @tpq__ on DeepTRIAGE, a machine learning tool for generating interpretable and individualised biomarker scores. It improved over other deep learning by providing easy to understand interpretations & also finds individualised variables to investigate #ABACBSGIW19 https://t.co/QiQitgdcwQ |
0.9933562 |
| CFlensburg | Elana Fertig @FertigLab kicking of the last day of #ABACBSGIW19 with “Transfer Learning for Cross-species, Cross-tissue, and Spatial Single Cell Analysis”. Starts by talking about different flavours of matrix factorisation. | 0.9926630 |
| samleechch | @MarkPinese predicting blood cancer using whole genome somatic cnv analysis for sub-clonal haematopoesis. Fascinating and well considered research. Survival plots pass the finger test #ABACBSGIW19 https://t.co/35gRGnAjxT | 0.9922591 |
| JovMaksimovic | The @abacbs postdoc survey is closing tomorrow at the end of the #ABACBSGIW19 conference. If you are a newbie postdoc (#combine19), a veteran postdoc or postdoc-identifying please complete the survey and let ABACBS know how they can best support and advocate for you. https://t.co/sIObXAFeEP | 0.9918083 |
| RuizCSergio | To conclude: we had a talk from UK via zoom, a talk from an iPad and also a talk with the voice inserted in the slides and played simultaneously. I love tech solutions, what a complete day!! #ABACBSGIW19 | 0.9918083 |
| PNgsabrina | It’s interesting to learn how dogs are highly similar to human in terms of their mental state 😯 Genomic research in dogs has helped us understand obsessive compulsive disorder in humans @abacbs #ABACBSGIW19 https://t.co/Je2tiuU3iQ | 0.9918083 |
| davisjmcc | Excellent talk this morning from @tpq__ at #ABACBSGIW19 - developing interpretable deep learning models by looking at the “attention” of models, in this case applied to breast cancer subtypes. Check out the preprint here: https://t.co/HXE6jNkoW2 | 0.9918083 |
Topic 4
| screen_name | text | gamma |
|---|---|---|
| shazanfar |
The most advanced artificial intelligence technique is still our eyeballs 👀👀 @trashystats presenting schex (pron ess-see-hex), a Bioconductor package to avoid overplotting of points so we can more effectively visualise single cell data #ABACBSGIW19 https://t.co/MloDm2TMS7 |
0.9926630 |
| methylnick | Great analogy. Using a lens of prior learned factors and look at orthogonal datasets. If biological the same cluster will be present. If technical, not so much. You could use this to back annotate clusters from original data. New tool projectR EF @abacbs #ABACBSGIW19 #singleCell https://t.co/7yKn72Fdx6 | 0.9926630 |
| methylnick | Located in a locus associated with OCD for working farm dogs. Kelpies. @abacbs #ABACBSGIW19 the descriptor maybe off? OCD traits aligned with farm dogs who are precise in sheep herding. Claire Wade. https://t.co/7j7lmtZPA7 | 0.9922591 |
| shazanfar |
Congrats to the excellent organisers of #combine19 & all @combine_au members & exec for a fantastic year of “for students, by students” events. 2020 exec nominations are now open, I found it incredibly rewarding & highly recommend! #ABACBSGIW19 @abacbs https://t.co/REEA4BjOuU |
0.9918083 |
| CFlensburg | David Lynn first plenary of the afternoon on “Rewiring of metabolic, transcriptional, and protein-protein interaction networks in colorectal cancer cells expressing oncogenic KRAS” #ABACBSGIW19 | 0.9918083 |
| abacbs | PW: Reiterate what Rafa discussed last night. Carefully look at your data. Batch correction and outlier detection important. Imputation from missing values and check sample labelling #ABACBSGIW19 | 0.9918083 |
| tsonika | Beautiful talk by @davisjmcc on population scale eQTL effect studies at single cell level. Preprint herehttps://www.biorxiv.org/content/10.1101/630996v1 #ABACBSGIW19 https://t.co/SSBmbtOT1Y | 0.9918083 |
| frostickle | Dogs are great to study because they’re widely available in Australia. Claire doesn’t actually keep dogs in her lab, people bring in their dogs for her to do cheek swabs on and then they go home #ABACBSGIW19 https://t.co/5sno7dcgNn | 0.9913016 |
| abacbs | DL: Interestingly, oncogenic KRAS cells are highly transcriptionally responsive to activation of EGFR via TGFa stimulation. Also a down-regulation of protein synthesis and metabolism #ABACBSGIW19 | 0.9913016 |
| methylnick | SC: introduces DELFI, DNA Evaluation of Fragments for early Interception. Low pass WGS on liquid biopsies. Looking at nucleosomal fragments. @abacbs #ABACBSGIW19 https://t.co/R4AVtVNywN | 0.9907282 |
Topic 5
| screen_name | text | gamma |
|---|---|---|
| frostickle |
Dr Kitty Lo presenting the Sierra R package, designed for detecting differential transcript usage analysis from polyA-enriched single cell RNASeq data. Check out their poster if you’re here at #ABACBSGIW19! Or their github here: https://t.co/lJPlGgTIy2 @AliciaOshlack https://t.co/spjdCwhCUq |
0.9939297 |
| DrSimonHo |
We seek a Research Assistant with a background in bioinformatics or computational biology to join our lab at @Sydney_Uni This is a fixed-term (1 year) position and will be advertised in January. For further info and contact details visit https://t.co/gshu1EBC5P #ABACBSGIW19 https://t.co/jEPLn2ty4J |
0.9933562 |
| JovMaksimovic | For everyone who will be attending the @abacbs post doc event at 6pm tonight in the beer garden at the Alfred Hotel (https://t.co/qzQ8iLSUtt) there will be a bar tab! Enjoy the 🍻 and say “Hi!” to Ignatius Pang who has kindly stepped in to host in my stead #combine19 #ABACBSGIW19 | 0.9933562 |
| frostickle | Double feature at #ABACBSGIW19 from @fredjaya1! Genetic population structure and reproductive character displacement within a cryptic species complex of Uperoleia toadlets and Assessing recombination detection methods using viral simulations https://t.co/PCHxOP1Equ | 0.9930268 |
| shazanfar | Kicking off the scientific program at #ABACBSGIW19 with @rafalab plenary talk on Statistical approaches for single-cell genomics. Workflows in statistics/bioinformatics are especially insidious, you always get an answer, even if there are glaring errors after first glance | 0.9926630 |
| paulfharrison |
Kitty Lo describing Sierra package for detecting differential transcripts (ie APA) from single cell data. One clever feature is a splice-aware peak caller. https://t.co/PkC1uFkwcM #ABACBSGIW19 |
0.9926630 |
| KelWyres | Enjoying #ABACBSGIW19 and want to pursue more exciting computational biology? We’re looking for a PhD student and a postdoc to work on new projects combining bacterial genomics and metabolic modeling. Check it out! PhD:https://t.co/4ydCJSmduD PostDoc:https://t.co/6cr8BCpBXa | 0.9926630 |
| methylnick | JM @MarioniLab taking us through batch effects in scRNASeq and how compositional effects confound batch, batch is something to be lived with given the technology. Using M&M approach to account, doing okay and better than other state of the art methods. @abacbs #ABACBSGIW19 https://t.co/gPVdDm8Aho | 0.9922591 |
| davisjmcc | So I’m sitting here quietly at #ABACBSGIW19 and at the start of her talk @trashystats accuse ME, your faithful correspondent, of spreading a false pronunciation of her “schex” package. Outrageous! For the record, it’s pronounced “shecks” | 0.9922591 |
| tsonika | .@EduEyras: RATTLE has the best precision in transcripts assemblies compared with similar tools. Less sensitivity ( missing some intron chains). Coming next - Differential splicing, RNA modifications, fusion etc #ABACBSGIW19 | 0.9918083 |
Topic 6
| screen_name | text | gamma |
|---|---|---|
| abacbs | DB: Beacon protocol for clinical variant sharing. The resource is unsustainable ($4K/month to maintain, 33 hours to update and 20sec query time). Serverless is cost effective. With sBeacon we’ve achieved a big cost saving and much quicker #ABACBSGIW19 | 0.9941808 |
| frostickle |
Introme: Identifying splice-altering variants as drivers of high-risk paediatric cancer, by Patricia Sullivan @PatSullivann from @UNSWScience Only 70% of cancer-causing mutations have drug treatments available, so we’re still relying on radiation & chemo. #ABACBSGIW19 https://t.co/8tKlvfRCZJ |
0.9936559 |
| CFlensburg | Qing Wang talking about variant calling from ion torrent data. Very low agreement between callsets from different methods. Especially mutect2 calls lots of variants. Consensus calling makes it more reliable. #ABACBSGIW19 | 0.9930268 |
| lazappi |
Time difference means no streaming for me but I can still stalk the #ABACBSGIW19 #ABACBSGIW2019 Twitter feed https://t.co/xc4TyU8qJE. Links to GitHub/Bitbucket/Bioconductor/CRAN will automatically join the list at the bottom so remember to tweet your (and others) work. |
0.9930268 |
| shazanfar | How do we optimise when we don’t necessarily have a reliable objective? USyd PhD student Tom Geddes describing use of Pareto Frontier to find the most stable/dominating set of parameter configurations for the datasets on hand #ABACBSGIW19 https://t.co/bGxicEBapD | 0.9930268 |
| shazanfar | #ABACBSGIW19 @davisjmcc presenting work together with @OliverStegle’s PhD student @AnnaCuomo92 on identifying expression quantitative trait loci (eQTL), i.e. genetic differences associated with gene expression, pointing towards potential important regulators of expression | 0.9922591 |
| frostickle |
Future work includes observing the dogs directly to validate what farmers have been saying about their dogs, and also reproducing the study in other breeds. Nevertheless, this is a very rich resource for studying the relationship between genes and cognition. #ABACBSGIW19 https://t.co/51ZGlrvIPK |
0.9922591 |
| markjcowley | Excellent talk today from @PatSullivann presenting her honours research: finding atypical splice altering variants in paediatric cancer via #introme. Representing BABS @UNSWScience and @KidsCancerInst. #ABACBSGIW19 https://t.co/QvDTVQVwUF | 0.9918083 |
| tpq__ |
Very honored to receive the “people’s choice award” for best talk at #ABACBSGIW19 – but must share the credit with my amazing team @adhambeyki @samleechch @truyenoz @DeakinA2I2 that made this work possible Check out our pre-print for the details 😁 https://t.co/A3bFEGsFzH |
0.9918083 |
| frostickle |
Congratulations to @NhiHin for giving the best talk of #COMBINE19 and being invited to present in front of everyone at #ABACBSGIW19 Nih’s work uses Zebrafish to model & investigate familial Alzheimer’s disease #Alzheimers @UniofAdelaide @UofABioinfoHub https://t.co/QDcX4eulg5 |
0.9913016 |
8 Links
Links to GitHub, GitLab, BitBucket, Bioconductor or CRAN mentioned in Tweets.
| Name | Tweets | Retweets | Type | Link |
|---|---|---|---|---|
| RATTLE | 2 | 5 | GitHub | comprna/rattle |
| scCCESS | 2 | 1 | GitHub | gedcom/scccess |
| gencore | 2 | 0 | GitHub | opengene/gencore |
| buscomp | 1 | 0 | GitHub | slimsuite/buscomp |
| FlowGrid | 1 | 0 | GitHub | holab-hku/flowgrid |
| projectR | 1 | 0 | GitHub | genesofeve/projectr |
| reorientexpress | 1 | 0 | GitHub | comprna/reorientexpress |
| scDECAF | 1 | 0 | GitHub | soroorh/scdecaf |
| Sierra | 1 | 0 | GitHub | vccri/sierra |
| VariantSpark | 1 | 0 | GitHub | aehrc/variantspark |
| XenoSplit | 1 | 0 | GitHub | goknurginer/xenosplit |
Session info
## ─ Session info ───────────────────────────────────────────────────────────────
## setting value
## version R version 4.0.0 (2020-04-24)
## os macOS Catalina 10.15.6
## system x86_64, darwin17.0
## ui X11
## language (EN)
## collate en_US.UTF-8
## ctype en_US.UTF-8
## tz Europe/Berlin
## date 2020-09-01
##
## ─ Packages ───────────────────────────────────────────────────────────────────
## package * version date lib source
## askpass 1.1 2019-01-13 [1] CRAN (R 4.0.0)
## assertthat 0.2.1 2019-03-21 [1] CRAN (R 4.0.0)
## backports 1.1.8 2020-06-17 [1] CRAN (R 4.0.0)
## bitops 1.0-6 2013-08-17 [1] CRAN (R 4.0.0)
## callr 3.4.3 2020-03-28 [1] CRAN (R 4.0.0)
## clamour * 0.1.0 2020-09-01 [1] Github (lazappi/clamour@c8ea1c7)
## cli 2.0.2 2020-02-28 [1] CRAN (R 4.0.0)
## colorspace 1.4-1 2019-03-18 [1] CRAN (R 4.0.0)
## crayon 1.3.4 2017-09-16 [1] CRAN (R 4.0.0)
## curl 4.3 2019-12-02 [1] CRAN (R 4.0.0)
## digest 0.6.25 2020-02-23 [1] CRAN (R 4.0.0)
## dplyr * 1.0.1 2020-07-31 [1] CRAN (R 4.0.2)
## ellipsis 0.3.1 2020-05-15 [1] CRAN (R 4.0.0)
## emo * 0.0.0.9000 2020-08-17 [1] Github (hadley/emo@3f03b11)
## evaluate 0.14 2019-05-28 [1] CRAN (R 4.0.0)
## fansi 0.4.1 2020-01-08 [1] CRAN (R 4.0.0)
## farver 2.0.3 2020-01-16 [1] CRAN (R 4.0.0)
## forcats * 0.5.0 2020-03-01 [1] CRAN (R 4.0.0)
## fs * 1.5.0 2020-07-31 [1] CRAN (R 4.0.2)
## generics 0.0.2 2018-11-29 [1] CRAN (R 4.0.0)
## ggforce 0.3.2 2020-06-23 [1] CRAN (R 4.0.2)
## ggplot2 * 3.3.2 2020-06-19 [1] CRAN (R 4.0.2)
## ggraph * 2.0.3 2020-05-20 [1] CRAN (R 4.0.0)
## ggrepel * 0.8.2 2020-03-08 [1] CRAN (R 4.0.0)
## ggtext * 0.1.0 2020-06-04 [1] CRAN (R 4.0.2)
## glue 1.4.1 2020-05-13 [1] CRAN (R 4.0.0)
## graphlayouts 0.7.0 2020-04-25 [1] CRAN (R 4.0.0)
## gridExtra 2.3 2017-09-09 [1] CRAN (R 4.0.0)
## gridtext 0.1.1 2020-02-24 [1] CRAN (R 4.0.2)
## gtable 0.3.0 2019-03-25 [1] CRAN (R 4.0.0)
## here * 0.1 2017-05-28 [1] CRAN (R 4.0.0)
## highr 0.8 2019-03-20 [1] CRAN (R 4.0.0)
## htmltools 0.5.0 2020-06-16 [1] CRAN (R 4.0.0)
## httr 1.4.2 2020-07-20 [1] CRAN (R 4.0.2)
## igraph * 1.2.5 2020-03-19 [1] CRAN (R 4.0.0)
## janeaustenr 0.1.5 2017-06-10 [1] CRAN (R 4.0.0)
## jsonlite 1.7.0 2020-06-25 [1] CRAN (R 4.0.0)
## kableExtra * 1.2.1 2020-08-27 [1] CRAN (R 4.0.2)
## knitr * 1.29 2020-06-23 [1] CRAN (R 4.0.0)
## labeling 0.3 2014-08-23 [1] CRAN (R 4.0.0)
## lattice 0.20-41 2020-04-02 [1] CRAN (R 4.0.0)
## lifecycle 0.2.0 2020-03-06 [1] CRAN (R 4.0.0)
## lubridate * 1.7.9 2020-06-08 [1] CRAN (R 4.0.0)
## magick * 2.4.0 2020-06-23 [1] CRAN (R 4.0.0)
## magrittr 1.5 2014-11-22 [1] CRAN (R 4.0.0)
## markdown 1.1 2019-08-07 [1] CRAN (R 4.0.0)
## MASS 7.3-51.6 2020-04-26 [1] CRAN (R 4.0.0)
## Matrix 1.2-18 2019-11-27 [1] CRAN (R 4.0.0)
## modeltools 0.2-23 2020-03-05 [1] CRAN (R 4.0.0)
## munsell 0.5.0 2018-06-12 [1] CRAN (R 4.0.0)
## NLP 0.2-0 2018-10-18 [1] CRAN (R 4.0.0)
## openssl 1.4.2 2020-06-27 [1] CRAN (R 4.0.0)
## pillar 1.4.6 2020-07-10 [1] CRAN (R 4.0.2)
## pkgconfig 2.0.3 2019-09-22 [1] CRAN (R 4.0.0)
## plyr 1.8.6 2020-03-03 [1] CRAN (R 4.0.0)
## png 0.1-7 2013-12-03 [1] CRAN (R 4.0.0)
## polyclip 1.10-0 2019-03-14 [1] CRAN (R 4.0.0)
## processx 3.4.3 2020-07-05 [1] CRAN (R 4.0.2)
## ps 1.3.3 2020-05-08 [1] CRAN (R 4.0.0)
## purrr * 0.3.4 2020-04-17 [1] CRAN (R 4.0.0)
## R6 2.4.1 2019-11-12 [1] CRAN (R 4.0.0)
## RColorBrewer * 1.1-2 2014-12-07 [1] CRAN (R 4.0.0)
## Rcpp 1.0.5 2020-07-06 [1] CRAN (R 4.0.0)
## RCurl 1.98-1.2 2020-04-18 [1] CRAN (R 4.0.0)
## reshape2 1.4.4 2020-04-09 [1] CRAN (R 4.0.0)
## rlang 0.4.7 2020-07-09 [1] CRAN (R 4.0.2)
## rmarkdown 2.3 2020-06-18 [1] CRAN (R 4.0.0)
## rprojroot 1.3-2 2018-01-03 [1] CRAN (R 4.0.0)
## rstudioapi 0.11 2020-02-07 [1] CRAN (R 4.0.0)
## rtweet * 0.7.0 2020-01-08 [1] CRAN (R 4.0.0)
## rvest * 0.3.6 2020-07-25 [1] CRAN (R 4.0.2)
## scales 1.1.1 2020-05-11 [1] CRAN (R 4.0.0)
## selectr 0.4-2 2019-11-20 [1] CRAN (R 4.0.0)
## sessioninfo 1.1.1 2018-11-05 [1] CRAN (R 4.0.0)
## slam 0.1-47 2019-12-21 [1] CRAN (R 4.0.0)
## SnowballC 0.7.0 2020-04-01 [1] CRAN (R 4.0.0)
## stringi 1.4.6 2020-02-17 [1] CRAN (R 4.0.0)
## stringr * 1.4.0 2019-02-10 [1] CRAN (R 4.0.0)
## tibble 3.0.3 2020-07-10 [1] CRAN (R 4.0.2)
## tidygraph 1.2.0 2020-05-12 [1] CRAN (R 4.0.0)
## tidyr * 1.1.1 2020-07-31 [1] CRAN (R 4.0.2)
## tidyselect 1.1.0 2020-05-11 [1] CRAN (R 4.0.0)
## tidytext * 0.2.5 2020-07-11 [1] CRAN (R 4.0.2)
## tm 0.7-7 2019-12-12 [1] CRAN (R 4.0.0)
## tokenizers 0.2.1 2018-03-29 [1] CRAN (R 4.0.0)
## topicmodels * 0.2-11 2020-04-19 [1] CRAN (R 4.0.0)
## tweenr 1.0.1 2018-12-14 [1] CRAN (R 4.0.0)
## usethis 1.6.1 2020-04-29 [1] CRAN (R 4.0.0)
## utf8 1.1.4 2018-05-24 [1] CRAN (R 4.0.0)
## vctrs 0.3.2 2020-07-15 [1] CRAN (R 4.0.2)
## viridis * 0.5.1 2018-03-29 [1] CRAN (R 4.0.0)
## viridisLite * 0.3.0 2018-02-01 [1] CRAN (R 4.0.0)
## webshot * 0.5.2 2019-11-22 [1] CRAN (R 4.0.0)
## withr 2.2.0 2020-04-20 [1] CRAN (R 4.0.0)
## wordcloud * 2.6 2018-08-24 [1] CRAN (R 4.0.0)
## xfun 0.16 2020-07-24 [1] CRAN (R 4.0.2)
## xml2 * 1.3.2 2020-04-23 [1] CRAN (R 4.0.0)
## yaml 2.2.1 2020-02-01 [1] CRAN (R 4.0.0)
##
## [1] /Library/Frameworks/R.framework/Versions/4.0/Resources/library